Take a look at the following file structure:
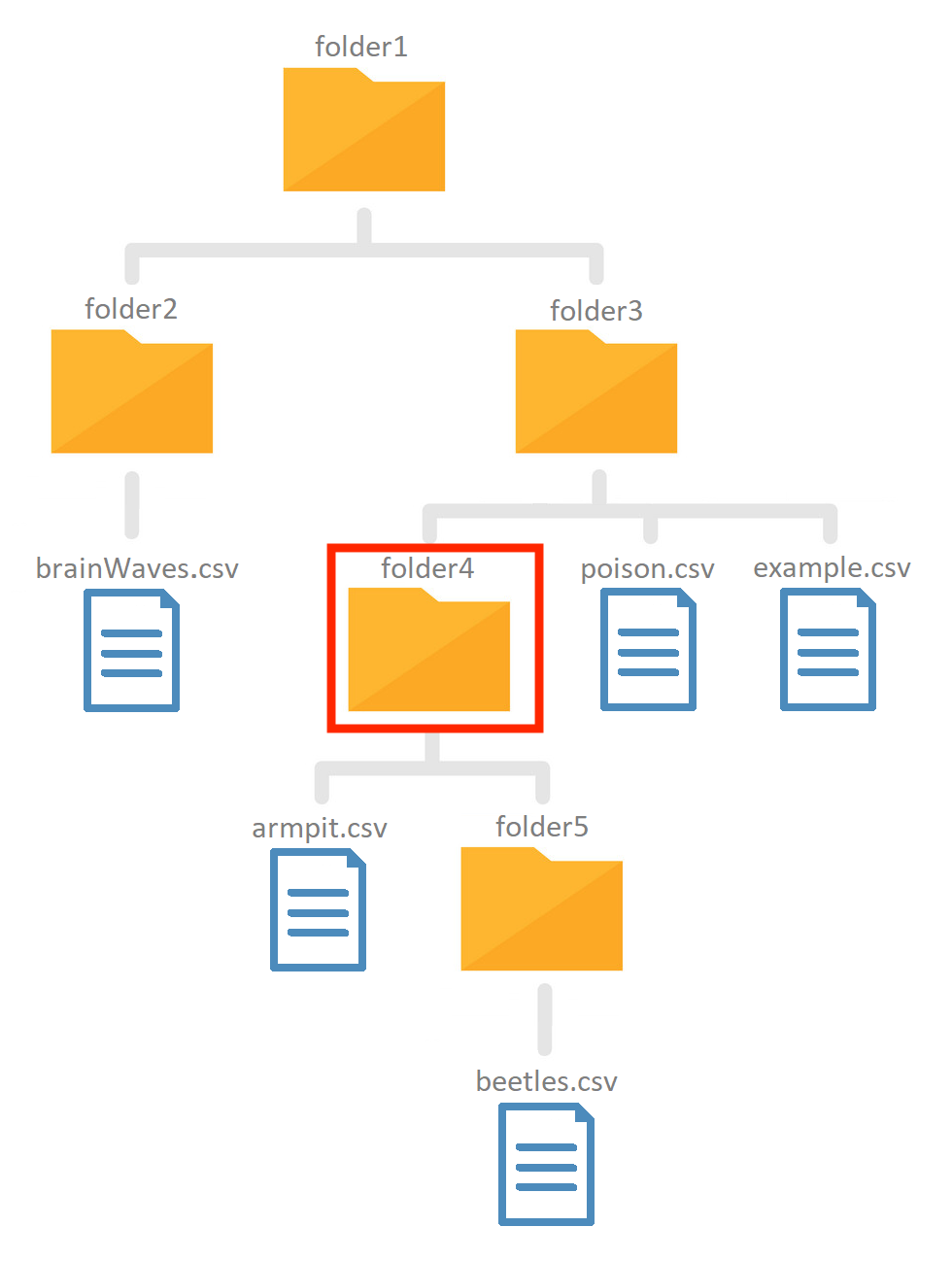
Note that this is only a small section of the total file structure. But when working with relative paths you dont even need to know the entire file structure.
Working in RStudio (For once not recommended)
If you want to make this exercise in RStudio you will have to download this zipped file structure1 and set your working directory to
folder4, if you place the zip-file in your working directory and unpack it you can use the following code to set the working directory:
setwd("folder1/folder3/folder4")
Exercise
!! Your current working directory is folder4, the red square !!. construct relative paths to the following files (based on the figure) and import them using the read_csv function from the readr package.
Example
- As an example we loaded in the
example.csvfile in theexampleobject. Because this file is located in a parent folder (folder3) of the working directory we need to instruct the pc to first ascent into this folder (using../):
example <- read_csv("../example.csv")
- In some cases the file will be located in a child folder and a similar path can be constructed (where we replaced
../withname_of_child_folder/):
read_csv("folder5/beetles.csv"). Folder5 is the child directory.
!! Your current working directory is folder4, the red square, look at the figure for the file structure !!
!! You can’t change your working directory !!
-
Load
poison.csvand store the dataframe inpoison. (Tip: you will need to use../) -
Load
armpit.csvand store the dataframe inarmpit. -
Load
beetles.csvand store the dataframe inbeetles. -
Load
brainWaves.csvand store the dataframe inbrainWaves. (Tip: you will need to use../)
Note: don’t forget to load the readr package and use its read_csv function, not the read.csv function.