In February 2013, brothers named Elwin and Yohan were arrested for six rapes in France, but both denied the charges. Deciding which is guilty is a tricky affair — they're identical twins, so the genetic difference between them is very slight. Marseille police chief Emmanual Kiehl said, "It could take thousands of separate tests before we know which one of them may be guilty."
This is only the latest in a series of legal conundrums involving identical twins and DNA evidence. During a jewel heist in Germany in January 2009, thieves left behind a drop of sweat on a latex glove. A crime database showed two hits — identical twins Hassan and Abbas O. (under German law their last name was withheld). Both brothers had criminal records for theft and fraud, but both were released. The court ruled, "From the evidence we have, we can deduce that at least one of the brothers took part in the crime, but it has not been possible to determine which one."
Later that year, identical twins Sathis Raj and Sabarish Raj escaped hanging in Malaysia when a judge ruled it was impossible to determine which was guilty of drug smuggling. "Although one of them must be called to enter a defence, I can't be calling the wrong twin to enter his defence," the judge told the court. "I also can't be sending the wrong person to the gallows."
In 2003, a Missouri woman had sex with identical twins Raymon and Richard Miller within hours of one another. When she became pregnant, both men denied fathering the child. In Missouri a man can be named a legal father only if a paternity test shows a 98 percent or higher probability of a DNA match, but the Miller twins both showed a probability of more than 99.9 percent.
"With identical twins, even if you sequenced their whole genome you wouldn't find difference," forensic scientist Bob Gaensslen told ABC News at the time. More recent research shows that this isn't the case, but teasing out the difference can be expensive — in the Marseilles case, police were told that such a test would cost €996,000.
It goes on. In August 2013, British authorities were trying to decide how to prosecute a rape when DNA evidence identified both Mohammed and Aftab Asghar. "It is an unusual case," said prosecutor Sandra Beck. "They are identical twins. The allegation is one of rape. There is further work due."
Assignment
Most differences found in the genomes of identical twins are due to copy-number variations (CNV). These structural variations alter the DNA of a genome such that cells have an abnormal or — for certain genes — a normal variation in the number of copies of one or more sections of the DNA. CNVs correspond to relatively large regions of the genome that have been removed (deletions) or duplicated (insertions) on certain chromosomes. For example, the chromosome that normally has sections in order as A-B-C-D might instead have sections A-B-C-C-C-D (a duplication of C) or A-B-D (a deletion of C).
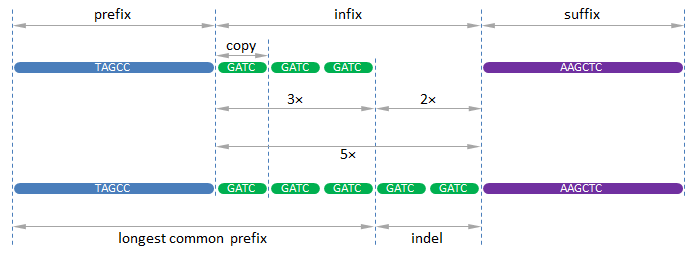
In order to detect CNVs, we assume a DNA sequence in composed as the concatenation of a prefix, followed by an infix and a suffix, where the infix is composed of $$n \in \mathbb{N}$$ repeats of a DNA-fragment that we call the copy.

In this assignment, we represent a DNA sequence as a string that only contains the uppercase letters A, C, G and T. Now, say that we have two DNA sequences that only differ in the number of repeats of the copy. Based on a comparison of both sequences, we can identify their different components. In order to do so, you proceed as follows:
-
Write a function replicate that has one mandatory parameter copy and three optional parameters number (default value: 1), prefix (default value: the empty string) and suffix (default value: the empty string). An positive integer must be passed to the the parameter number, and a DNA sequence must be passed to the other parameters. The function must return the DNA sequence that if composed as the concatenation of the given prefix, followed by an infix and the given suffix, where the infix is composed of the given number of repeats of the given copy.
-
Write a function copy_number that takes a DNA sequence $$s$$. The function must return a tuple that contains a DNA sequence $$c$$ and an integer $$n \in \mathbb{N}_0$$, where $$c$$ is the shortest possible DNA sequence for which the given DNA sequence $$s$$ is composed out of $$n$$ repeats of $$k$$.
-
Write a function LGE that takes two DNA sequences. The function also has an optional parameter suffix (default value: False) that takes a Boolean value. If the value False is passed to the parameter suffix, the function must return the longest common prefix (longest common string at the start of the given sequences). If the value True is passed to the parameter suffix, the function must return the longest common suffix (longest common string at the end of the given sequences).
-
Now, use the previous two functions to write a function CNV that takes two DNA sequences. In case both DNA sequences have the same prefix and suffix, and only differ in the number of intermediate repeats of the same copy, the function must return a tuple that contains the (shortest possible) copy and the number of repeats of this copy in the first and in the second sequence. Otherwise, the function must raise an AssertionError with the message no CNV found. Use the following procedure to identify the different components in the two given DNA sequences (see figure below):
-
check that the sequences differ, otherwise no CNV is found
-
determine the longest common prefix (LCP) of the sequences
-
determine the suffix as the remaining part of the shortest sequence (tail not belonging to the LCP)
-
check that the longest sequence ends with the suffix found, otherwise no CNV is found
-
determine the indel as the part in between the LCP and the suffix of the longest sequence
-
determine the shortest possible copy and the number of repeats of that copy from which the indel is composed (in case of CNV the indel is composed of one or more repeats of the copy)
-
determine the number of extra repeats of the copy at the end of the LCP
-
Example
>>> replicate('GATC')
'GATC'
>>> replicate('GATC', number=4)
'GATCGATCGATCGATC'
>>> replicate('GATC', number=2, prefix='TAGCC')
'TAGCCGATCGATC'
>>> replicate(copy='GATC', number=3, suffix='AAGCTC')
'GATCGATCGATCAAGCTC'
>>> replicate(copy='GATC', number=3, prefix='TAGCC', suffix='AAGCTC')
'TAGCCGATCGATCGATCAAGCTC'
>>> replicate(copy='GATC', number=5, suffix='AAGCTC', prefix='TAGCC')
'TAGCCGATCGATCGATCGATCGATCAAGCTC'
>>> copy_number('CTCTCTCTCTCTCTCTCTCTCTCT') # replicate(copy='CT', number=12)
('CT', 12)
>>> copy_number('GATCGATCGATCGATC') # replicate(copy='GATC', number=4)
('GATC', 4)
>>> copy_number(replicate('GATCGATC', number=2))
('GATC', 4)
>>> copy_number('GATCGATCGATCGATCG') # replicate(copy='GATC', number=4) + 'G'
('GATCGATCGATCGATCG', 1)
>>> seq1 = 'TAGCCGATCGATCGATCAAGCTC' # replicate(copy='GATC', number=3, prefix='TAGCC', suffix='AAGCTC')
>>> seq2 = 'TAGCCGATCGATCGATCGATCGATCAAGCTC' # replicate(copy='GATC', number=5, suffix='AAGCTC', prefix='TAGCC')
>>> LCE(seq1, seq2)
'TAGCCGATCGATCGATC'
>>> LCE(seq1, seq2, suffix=True)
'CGATCGATCGATCAAGCTC'
>>> LCE(seq1, seq1)
'TAGCCGATCGATCGATCAAGCTC'
>>> seq1 = 'TAGCCGATCGATCGATCAAGCTC' # replicate(copy='GATC', number=3, prefix='TAGCC', suffix='AAGCTC')
>>> seq2 = 'TAGCCGATCGATCGATCGATCGATCAAGCTC' # replicate(copy='GATC', number=5, suffix='AAGCTC', prefix='TAGCC')
>>> CNV(seq1, seq2)
('GATC', 3, 5)
>>> CNV(seq2, seq1)
('GATC', 5, 3)
>>> CNV(seq1, seq1)
Traceback (most recent call last):
AssertionError: no CNV found
>>> seq3 = 'TAGCGATCGATCGATCGATCGATCAAGCTC' # replicate(copy='GATC', number=5, prefix='TAGC', suffix='AAGCTC')
>>> CNV(seq1, seq3)
('GATCGAT', 0, 1)
>>> seq4 = 'TAGCCGATCGATCGATCGATCGATCAGCTC' # replicate(copy='GATC', number=5, suffix='AGCTC', prefix='TAGCC')
>>> CNV(seq1, seq4)
Traceback (most recent call last):
AssertionError: no CNV found