In bioinformatics, FASTA format is a text-based format for representing either nucleotide sequences or peptide sequences, in which nucleotides or amino acids are represented using single-letter codes. The format originates from the FASTA software package, but has now become a de facto standard in the field of bioinformatics. FASTA files may contain multiple records, where each record corresponds to a single nucleotide or amino acid sequence. A record in FASTA format begins with a single-line description, followed by lines of sequence data. The description line is distinguished from the sequence data by a greater-than (>) symbol in the first column.
The example FASTA file given below contains four records. In order to point out that the number of sequence characters per line is not fixed, we have split the sequences in the example over several lines with a different number of characters for each of the records. But even within a single record it is allowed to have sequence fragments of variable length.
>read1
AAAAAAAAGGTGCGGAGGAAAGCAACACATTTGTTCCTCAGGACTCTTCA
GCGGGAGATATCTGCAGAACCAAACACGCTCAAAGACCCGCGCAAATCGG
CAAATTGCCTGACGTAGAACACCGACCTAGCGTGTTTATTATGATACTCG
GCACCTCTGACTTAATCAAACGTTGTCGAGGTGGAGATGGTATCATCTGG
CGTTAGGCATAACGAGCGTGACACTAGCTTC
>read2
CTCTCGGAAGTTTGTCCGCACCGACATAAATAGACTGATACTGATCAGGGGGACGGTACG
ACCCACTCTGTCGATCGAACCAGTGACCTGTTCGCTTCGTAACTGGCCAGACGATAGATC
TTAGCATAACCGACGCGAAGTGTGGCAGATAAGATCCCAAGGTAGTAAATAGTACATATT
AGTGGTCAACAGGTTTTTAGCGCAGCAGCTGATCTATGCAATTGACTGCAACACCATGAC
GTAGGTTGCTTCTATAAGAACAAGTTTACCGCTACGAATTCGCGTCGGTTCCGTACATA
>read3
AAAAAAAAACGTCAGAAACGTGAGTAGGGTTACCCACACGCATCTAAGAATGCTCGGGCAATGTGACGGT
ATGAAGTTGAGAGCCTCTGTTACCCTCCACCTGATGGGGTGGCGGCCATTTACAGTATTGCTTAGCGCAC
TCAGATATACGCATGATGGGACTGATTCCCCAGGCGAGTACGTAGTCACCCGCGGCGACTCGACAAGGAT
ACTATTATCAGGGTTCTCCCCGGGAGGAGGTATTAAGA
>read4
AAAAAAAAAATGTCATGACCCTAGAAGGCCCTGCATATACATGGCAGGGCAGTCTATCAGCGCCCATCATCATCGCTGAC
GTAGTTGGAGCCGTATCTGTACTGGATCTAGGGGGCATCGTGAACTAGCGAGGTCGTGTGACGCGCTACAAAGGCTCGGC
CCATCTGGAACGTCAGACGAGGCTTCTCTACGGGGGTCCTGCCGTGGGCTATTGAGGTGCAAAGTTCGAATTCGGCACTG
TCGCGTGTAATTGAATTCGTGCCCAGT
Determining an organism's complete genome (called genome sequencing) forms a central task of bioinformatics. Unfortunately, we still don't possess the microscope technology to zoom into the nucleotide level and determine the sequence of a genome's nucleotides, one at a time. However, researchers can apply chemical methods to generate and identify much smaller snippets of DNA, called reads.
Because the current generation of sequencing machines produce so many reads at once, it is possible to sequence different organisms in parallel. This is done by fragmenting the DNA of the organisms, and sequencing these fragments in a single run. In order to link the sequenced fragments to the organism they originate from, each of the fragments is labeled with some kind of an organism-specific barcode. 454 Life Sciences (Roche) sequencers use a barcode of eight nucleotides that prefix each of the sequence fragments.
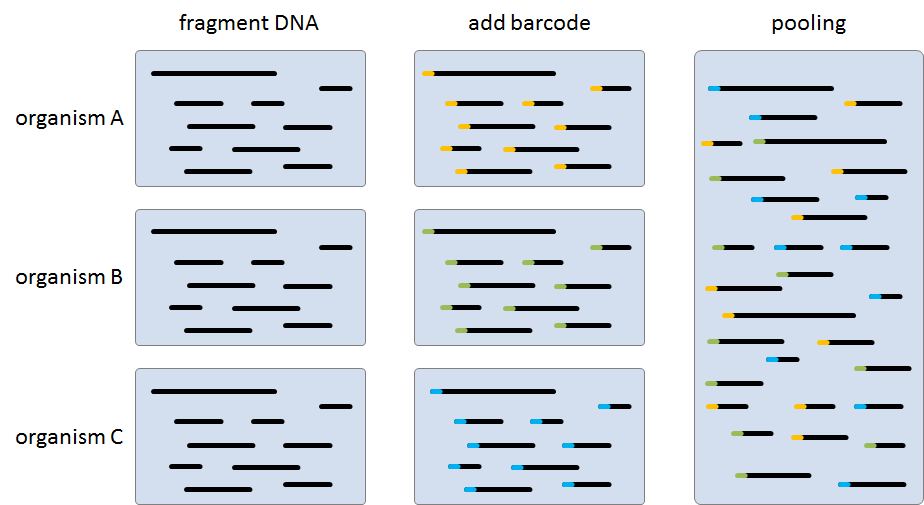
Assignment
You are given a FASTA file containing the reads generated by a single multiplexed run on a 454 Life Sciences sequencer. Your task is to split this file into multiple FASTA files, each containing the reads grouped per label. This is done in the following way:
Implement a function outputFasta that can be used to output a single sequence in FASTA format. The function takes two string arguments: a description and the sequence itself. In generating the output, the function must fragment the sequence into fixed-length strings (except for the last fragment). The default length is 80 characters, but specific lengths can be passed to the optional argument width. By default, the function prints out the sequence in FASTA format. However, if a file object that is opened for writing is passed to the optional argument file, the function instead must write the FASTA formatted sequence to this file. Make sure that each line written by the function, including the last line, ends with a newline ('\n').
Use the function outputFasta to implement a function demultiplexFasta. The function demultiplexFasta takes as its argument the location of a FASTA file containing a series of reads generated by a single multiplexed run on a 454 Life Sciences sequencer. The function must write all FASTA records from the given file that have the label XXXXXXXX to the file XXXXXXXX.fasta (existing files must be overwritten). In doing so, the order of the records in the given FASTA file must be retained. This procedure must be executed for all labels that prefix the reads in the given file. In writing FASTA records to the new files, the function must remove the labels from the sequences and must split the sequences into fixed-length fragments. The default length is 80 characters, but specific lengths can be passed to the optional argument width.
Example
In the following interactive session we assume the current directory to contain the text file reads.fasta. The content of this file is the same as the example FASTA file displayed in the introduction.
>>> outputFasta('read1', 'GGTGCGGAGGAAAGCAACACATTTGT', width=10)
>read1
GGTGCGGAGG
AAAGCAACAC
ATTTGT
>>> output = open('out.fasta', 'w')
>>> outputFasta('read2', 'AGTTTGTCCGCACCGACATAAATAGA', width=10, file=output)
>>> outputFasta('read3', 'ACGTCAGAAACGTGAG', width=10, file=output)
>>> output.close()
>>> print(open('out.fasta', 'r').read(), end='')
>read2
AGTTTGTCCG
CACCGACATA
AATAGA
>read3
ACGTCAGAAA
CGTGAG
>>> demultiplexFasta('reads.fasta', width=60)
>>> print(open('AAAAAAAA.fasta', 'r').read(), end='')
>read1
GGTGCGGAGGAAAGCAACACATTTGTTCCTCAGGACTCTTCAGCGGGAGATATCTGCAGA
ACCAAACACGCTCAAAGACCCGCGCAAATCGGCAAATTGCCTGACGTAGAACACCGACCT
AGCGTGTTTATTATGATACTCGGCACCTCTGACTTAATCAAACGTTGTCGAGGTGGAGAT
GGTATCATCTGGCGTTAGGCATAACGAGCGTGACACTAGCTTC
>read3
ACGTCAGAAACGTGAGTAGGGTTACCCACACGCATCTAAGAATGCTCGGGCAATGTGACG
GTATGAAGTTGAGAGCCTCTGTTACCCTCCACCTGATGGGGTGGCGGCCATTTACAGTAT
TGCTTAGCGCACTCAGATATACGCATGATGGGACTGATTCCCCAGGCGAGTACGTAGTCA
CCCGCGGCGACTCGACAAGGATACTATTATCAGGGTTCTCCCCGGGAGGAGGTATTAAGA
>read4
AATGTCATGACCCTAGAAGGCCCTGCATATACATGGCAGGGCAGTCTATCAGCGCCCATC
ATCATCGCTGACGTAGTTGGAGCCGTATCTGTACTGGATCTAGGGGGCATCGTGAACTAG
CGAGGTCGTGTGACGCGCTACAAAGGCTCGGCCCATCTGGAACGTCAGACGAGGCTTCTC
TACGGGGGTCCTGCCGTGGGCTATTGAGGTGCAAAGTTCGAATTCGGCACTGTCGCGTGT
AATTGAATTCGTGCCCAGT
>>> print(open('CTCTCGGA.fasta', 'r').read(), end='')
>read2
AGTTTGTCCGCACCGACATAAATAGACTGATACTGATCAGGGGGACGGTACGACCCACTC
TGTCGATCGAACCAGTGACCTGTTCGCTTCGTAACTGGCCAGACGATAGATCTTAGCATA
ACCGACGCGAAGTGTGGCAGATAAGATCCCAAGGTAGTAAATAGTACATATTAGTGGTCA
ACAGGTTTTTAGCGCAGCAGCTGATCTATGCAATTGACTGCAACACCATGACGTAGGTTG
CTTCTATAAGAACAAGTTTACCGCTACGAATTCGCGTCGGTTCCGTACATA