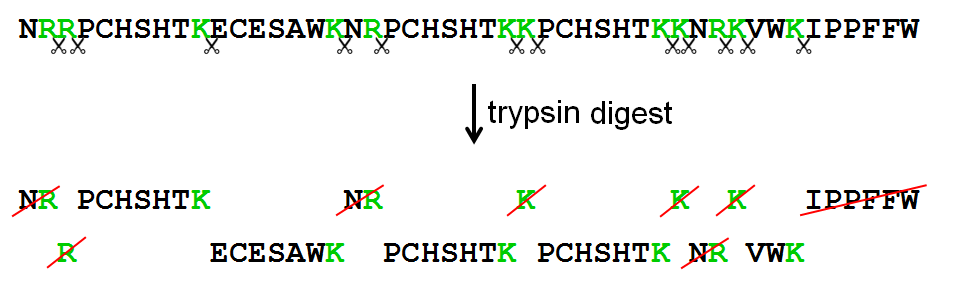
In this exercise we represent protein sequences as strings that only contain upper case letters. Each letter represents an amino acid within the protein sequence. Trypsin is a serine protease found in the digestive system of humans and many other vertebrates, where it helps to digest food proteins. The enzyme has a very specific function — it only cleaves peptide chains at the carboxyl side of the amino acids lysine (represented by the letter K) or arginine (represented by the letter R). As such, it is often used in laboratories studying protein structures.
High-performance liquid chromatography (HPLC) is a chromatographic technique used to separate the components in a mixture, to identify each component, and to quantify each component. When combined with shotgun tandem mass spectrometric methods, the active proteins within a biological sample may be determined. A trypsin digest is used to cleave the proteins in a sample downstream to every K or R. The individual components that result after the cleavage step are called tryptic peptides. The amino acid sequence of these tryptic peptides may then be determined by means of mass spectrometry. However, most devices have a detection limit that only allows to determine the amino acid sequence of peptides having a length between 5 and 50 amino acids. Further, if the last peptide of the protein chain does not end with K or R, it will not be picked up by the mass spectrometer.
Software suites such as Unipept are based on large protein databases, containing tryptic peptides taken from more than 29 million known proteins. This online platform can be used to determine both the diversity and the functional activity of a biological sample by comparing the tryptic peptides found in the sample with those found in the database.
Assignment
Define a class proteinDB that can be used to create simple protein databases. These protein databases can be used to look for proteins that contain a given list of tryptic peptides. The objects of the class proteinDB must support at least the following methods:
An initialization method that assures that each newly created object of the class proteinDB has a property peptides that refers to a dictionary. This dictionary should initially be empty for newly created objects, but gradually it will be filled with strings as keys and sets of strings as values.
A method addPeptide that can be used to add a new tryptic peptide to the database. The method takes two arguments: a string containing the label of a protein sequence and a string containing a tryptic peptide. Only tryptic peptides having a length between 5 and 50 residues (including boundaries) may be added to the database. In addition, tryptic peptides should end with K or R, and no other K or R may occur within the sequence. The method should throw an AssertionError with the message invalid peptide if a peptide is passed that does not meet all these conditions. Adding a tryptic peptide to the database is done by using the given tryptic peptide as a key in the dictionary that is referred to by the property peptides, and adding the given label of the protein sequences to the set that is mapped to this key by the dictionary. If the dictionary has no key for the given tryptic peptide, a new key-value pair must be added to the dictionary, with the peptide as the key and as its corresponding value a set that only contains the given label.
A method addProtein that can be used to add all tryptic peptides of a given protein that have a length between 5 and 50 (including boundaries) to the database. Two arguments must be passed to this method: a string containing the label of the protein sequence and a string containing the protein sequence itself. The method must perform an in silico tryptic digest to cut the protein sequences into its tryptic peptides, and add each of these tryptic peptides to the database under the label of the protein sequence.
A method addProteins that takes the location of a text file as an argument. Each line of this text file must contain the label of a protein sequence and the protein sequence itself, separated by a tab. The method must add all peptides of all proteins to the database.
A method identify that can be used to identify proteins from the database. This method takes a collection object (e.g. a list, tuple, set, …) that contains a number of peptides. The method must return an alphabetically ordered list of the labels of all proteins from the database that contain each of the given peptides at least once.
Take care in implementing these methods that you make optimal reuse of the methods that have already been implemented.
Example
The the following interactive session we assume that the text file proteins.txt is located in the current directory.
>>> unipept = proteinDB()
>>> unipept.addPeptide('PROT0001', 'ECESAWK')
>>> unipept.peptides
{'ECESAWK': {'PROT0001'}}
>>> unipept.addPeptide('PROT0002', 'WHK')
Traceback (most recent call last):
AssertionError: invalid peptide
>>> unipept.addPeptide('PROT0002', 'ESHLSTLAVQENEIG')
Traceback (most recent call last):
AssertionError: invalid peptide
>>> unipept.addPeptide('PROT0002', 'NWAQNAKIGGADWDCVCR')
Traceback (most recent call last):
AssertionError: invalid peptide
>>> unipept.addProtein('PROT0002', 'HAEWTDNQCCPVLKECESAWKYEMWQHPGEQHKRRRYEMWQHPGEQHKPCHSHTKVWKRY')
>>> unipept.peptides
{'ECESAWK': {'PROT0002', 'PROT0001'}, 'PCHSHTK': {'PROT0002'}, 'HAEWTDNQCCPVLK': {'PROT0002'}, 'YEMWQHPGEQHK': {'PROT0002'}}
>>> unipept.addProtein('PROT0003', 'NRRPCHSHTKECESAWKNRPCHSHTKKPCHSHTKKNRKVWKIPPFFW')
>>> unipept.peptides
{'ECESAWK': {'PROT0003', 'PROT0002', 'PROT0001'}, 'PCHSHTK': {'PROT0003', 'PROT0002'}, 'HAEWTDNQCCPVLK': {'PROT0002'}, 'YEMWQHPGEQHK': {'PROT0002'}}
>>> unipept.addProtein('PROT0004', 'YEMWQHPGEQHKECESAWKVPYCGFITRPCHSHTKECESAWK')
>>> unipept.peptides
{'ECESAWK': {'PROT0004', 'PROT0003', 'PROT0002', 'PROT0001'}, 'PCHSHTK': {'PROT0004', 'PROT0003', 'PROT0002'}, 'HAEWTDNQCCPVLK': {'PROT0002'}, 'VPYCGFITR': {'PROT0004'}, 'YEMWQHPGEQHK': {'PROT0004', 'PROT0002'}}
>>> unipept.identify(['VPYCGFITR'])
['PROT0004']
>>> unipept.identify({'ECESAWK', 'PCHSHTK'})
['PROT0002', 'PROT0003', 'PROT0004']
>>> unipept.identify(('YEMWQHPGEQHK', 'ECESAWK', 'PCHSHTK'))
['PROT0002', 'PROT0004']
>>> unipept.identify({'PCHSHTK', 'VPYCGFITR'})
['PROT0004']
>>> unipept.addProteins('proteins.txt')
>>> unipept.peptides
{'ECESAWK': {'PROT0005', 'PROT0004', 'PROT0003', 'PROT0002', 'PROT0001'}, 'VCEFPWFPMLINDVCR': {'PROT0007'}, 'VPYCGFITR': {'PROT0005', 'PROT0004'}, 'YEMWQHPGEQHK': {'PROT0006', 'PROT0005', 'PROT0004', 'PROT0002'}, 'PCHSHTK': {'PROT0006', 'PROT0005', 'PROT0004', 'PROT0003', 'PROT0002'}, 'HAEWTDNQCCPVLK': {'PROT0002'}, 'CSFHCLEK': {'PROT0006'}, 'AFNYMMPNTK': {'PROT0006'}, 'AYDDEVASFPGCMMATK': {'PROT0007', 'PROT0006'}, 'FIPYYPIYSR': {'PROT0006'}, 'TLCHETMR': {'PROT0005'}, 'HTPNYGVMWMFMNEWMSYDR': {'PROT0006', 'PROT0005'}, 'CDQMHVFDIYMIAIACSWGGPPSLTK': {'PROT0007'}, 'FGHSMTR': {'PROT0005'}}
>>> unipept.identify(('YEMWQHPGEQHK', 'VPYCGFITR', 'ECESAWK'))
['PROT0004', 'PROT0005']
>>> unipept.identify(('PCHSHTK', 'AYDDEVASFPGCMMATK'))
['PROT0006']
>>> unipept.identify(['NEGNLNVMK'])
[]