Here we use the gbm package, and within it the gbm() function, to fit boosted
regression trees to the Boston data set. We run gbm() with the option
distribution="gaussian" since this is a regression problem; if it were a binary
classification problem, we would use distribution="bernoulli". The
argument n.trees=5000 indicates that we want 5000 trees, and the option
interaction.depth=4 limits the depth of each tree.
library(gbm)
set.seed(1)
boost.boston <- gbm(medv ~ ., data = Boston[train,], distribution = "gaussian", n.trees = 5000, interaction.depth = 4)
The summary() function produces a relative influence plot and also outputs
the relative influence statistics.
summary(boost.boston)
var rel.inf
rm rm 43.9919329
lstat lstat 33.1216941
crim crim 4.2604167
dis dis 4.0111090
nox nox 3.4353017
black black 2.8267554
age age 2.6113938
ptratio ptratio 2.5403035
tax tax 1.4565654
indus indus 0.8008740
rad rad 0.6546400
zn zn 0.1446149
chas chas 0.1443986
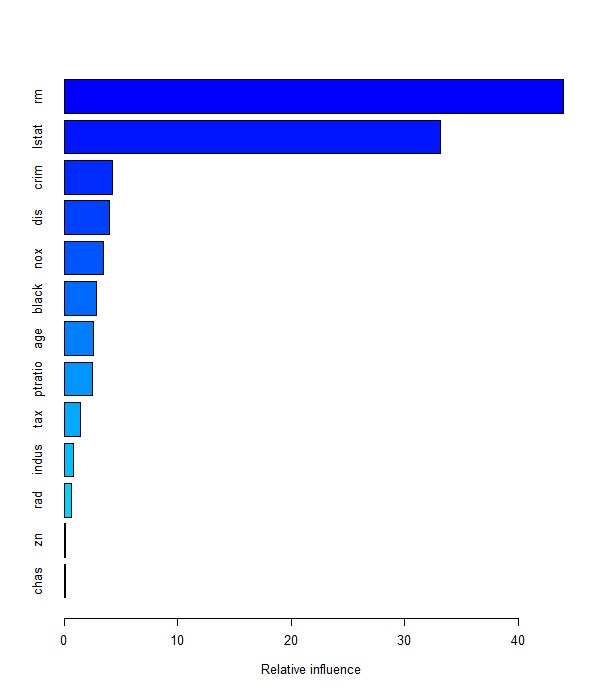
We see that lstat and rm are by far the most important variables. We can
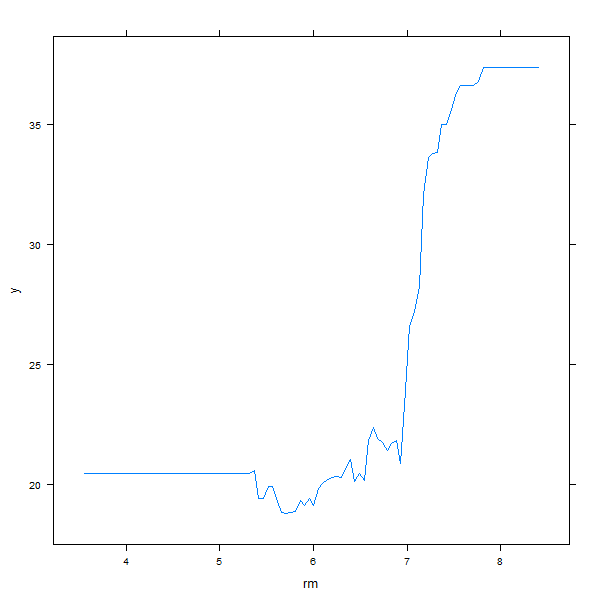
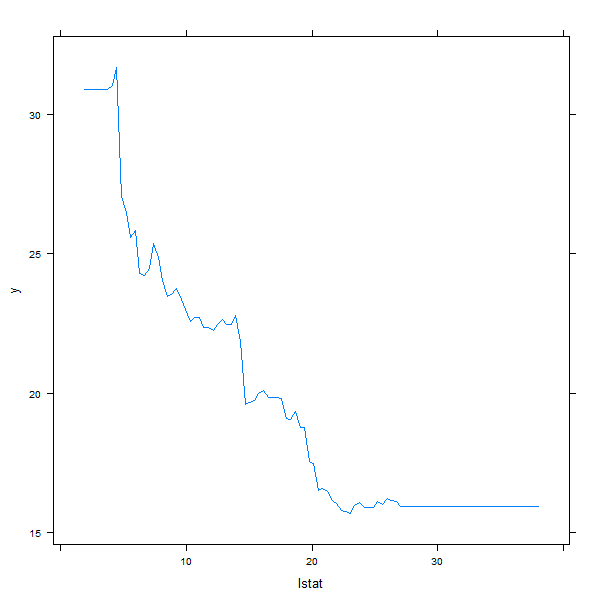
also produce partial dependence plots for these two variables. These plots
illustrate the marginal effect of the selected variables on the response after
integrating out the other variables. In this case, as we might expect, median
house prices are increasing with rm and decreasing with lstat.
plot(boost.boston, i = "rm")
plot(boost.boston, i = "lstat")
We now use the boosted model to predict medv on the test set:
yhat.boost <- predict(boost.boston, newdata = Boston[-train,], n.trees = 5000)
mean((yhat.boost - boston.test)^2)
[1] 18.84709
The test MSE obtained is 18.85; similar to the test MSE for random forests and superior to that for bagging. If we want to, we can perform boosting with a different value of the shrinkage parameter \(\lambda\) in \(\hat f(x) \leftarrow \hat f(x) + \lambda \hat f^b(x)\). The default value is 0.1, but this is easily modified. Here we take \(\lambda = 0.2\).
boost.boston <- gbm(medv ~ ., data = Boston[train,], distribution = "gaussian", n.trees = 5000, interaction.depth = 4, shrinkage = 0.2, verbose = F)
yhat.boost <- predict(boost.boston, newdata = Boston[-train,], n.trees = 5000)
mean((yhat.boost - boston.test)^2)
[1] 18.33455
In this case, using \(\lambda = 0.2\) leads to a slightly lower test MSE than \(\lambda = 0.1\).
Questions
The Smarket data set contains the daily percentage returns for the S&P 500 stock index between 2001 and 2005.
In this question we will try to predict whether a stock will go up or down (Direction) based on the its activity of the previous 5 days (Lag1, Lag2…).
library(ISLR2)
data(Smarket)
dim(Smarket)
[1] 1250 9
head(Smarket)
Year Lag1 Lag2 Lag3 Lag4 Lag5 Volume Today Direction
1 2001 0.381 -0.192 -2.624 -1.055 5.010 1.1913 0.959 Up
2 2001 0.959 0.381 -0.192 -2.624 -1.055 1.2965 1.032 Up
3 2001 1.032 0.959 0.381 -0.192 -2.624 1.4112 -0.623 Down
4 2001 -0.623 1.032 0.959 0.381 -0.192 1.2760 0.614 Up
5 2001 0.614 -0.623 1.032 0.959 0.381 1.2057 0.213 Up
6 2001 0.213 0.614 -0.623 1.032 0.959 1.3491 1.392 Up
- Create a new column
Targetin theSmarketdataframe which has the value 1 if the stock went up and 0 if the stock went down. - Create a training set of all the observations from 2001 to 2004 (2004 included). Store the dataframe in
train. -
Create a test set of all the observations in 2005. Store the dataframe in
test. - Train a boosted classification tree on the training data with
Targetas the dependent variable andLag1toLag5as the independent variables and store in an object namedfit.- For the individual classifiers use 100 decision stumps, these are decision trees with a depth of 1
- Set the shrinkage parameter to 0.2
- Using a threshold of 0, calculate the predicted value on the test data and store them in
preds. (If the model predicts a value smaller than 0, output 0, if the model predicts a value higher or equal to 0, output 1)
MC1: Does the model perform better than random guessing (accuracy higher than 50%)?
- 1: Yes
- 2: No
Assume that:
- The gbm and ISLR2 libraries have been loaded
- The Smarket data set has been loaded and attached