The FASTQ format is a text-based format for storing both DNA-sequences and their corresponding quality scores. It was originally developed at the Wellcome Trust Sanger Institute, but has recently become the de facto standard for storing the output of high throughput sequencing instruments.
A FASTQ file contains a series of sequence records, with each record corresponding to four consecutive lines:
-
line 1 begins with the at-sign (@) and is followed by a sequence identifier and an optional description
-
line 2 contains a series of sequence letters
-
line 3 begins with the plus-sign (+) and is optionally followed by the same sequence identifier and any description
-
line 4 encodes the quality values for the sequence in line 2, and must contain the same number of symbols as there are letters in the sequence
Both the sequence letters and quality scores are encoded with a single ASCII character for brevity. As an example, the content of the FASTQ file sample.fq containing six sequence records looks like this:
@sample sequence 1
GGAAGTCCATGGAGTTATTTGCGGTAACCGGGACCGGCCT
+sample sequence 1
ba^aaabaa`]baaaaa_aab]D^^`b`aYDW]abaa`^a
@sample sequence 2
CTCCTTATAGCATCATACGCACAGCTGGTATCCCCATTGCTTATTAGGGTTCAGCCCTGTCGCTCTCCCC
+sample sequence 2
ababa``aaaababaaaabaa_``aa``]^aa_`aa_Z_aaaaa^baaaaaaaaaaaaaaaaaa`^```a
@sample sequence 3
GGAAGTCCATGGAGTTATTTGCGGTAACCGGGACCGGCCTGGGGTTAGATTTTATGTCGT
+sample sequence 3
ba^aaabaa`]baaaaa_aab]D^^`b`aYDW]abaa`^ababbaaabaaaaa`]`ba`]
@sample sequence 4
TATAC
+sample sequence 4
baab`
@sample sequence 5
CTCCTTATAGCATCATACGCACAGCTGGTATCCCCATTGCTTATTAGGGT
+sample sequence 5
ababa``aaaababaaaabaa_``aa``]^aa_`aa_Z_aaaaa^baaaa
@sample sequence 6
GGAAGTCCATGGAGTTATTTGCGGTAACCGGGACCGGCCTGGGGTTAGATTTTATGTCGT
+sample sequence 6
ba^aaabaa`]baaaaa_aab]D^^`b`aYDW]abaa`^ababbaaabaaaaa`]`ba`]Prior to sequencing itself, a genomic library has to be constructed. As an artifact of the PCR amplification during library preparation, the same fragment may be read multiple times. That's why the DNA sequences in a FASTQ file are usually deduplicated, in order to avoid a possible bias when analyzing the sequences. Deduplication does not take into account full length sequences, but only relies on a prefix formed by the first $$n$$ bases of the sequence.
For example, if we consider prefixes of length 10, records 1, 3 and 6 in the sample FASTQ file share the same prefix GGAAGTCCAT. Similarly, records 2 and 4 also share the same prefix CTCCTTATAG. We have graphically depicted the records of this FASTQ file below, where the same prefixes have been given the same color.
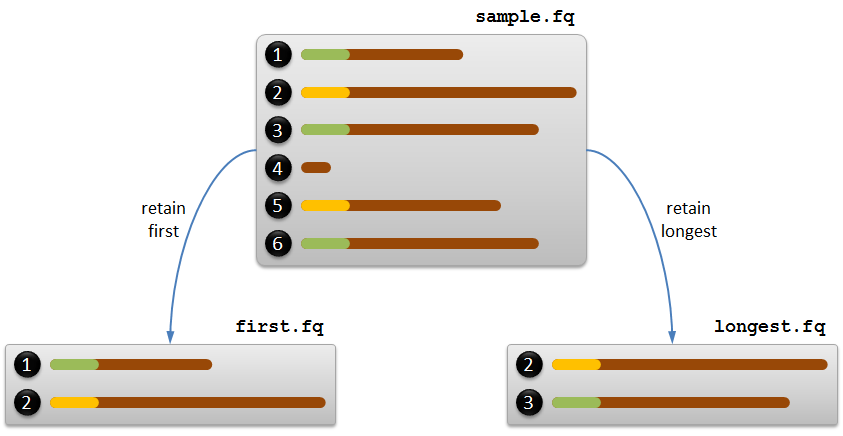
In practice, two strategies are used for deduplicating the sequences in a FASTQ file. Both strategies have in common that all records whose sequence is shorter than $$n$$ bases (the length of the prefix) are always removed. If $$n = 10$$, this is for example the case for record 4 in the sample FASTQ file.
In the first strategy — resulting in the left outcome show in the above figure — a FASTQ record is only retained if the prefix of its sequence is seen for the first time. To implement this strategy, the FASTQ records are read one by one and a set of prefixes is constructed to remember which prefixes have already been seen. A FASTQ record is then only retained in case its prefix was not yet in the set.
In the second strategy — resulting in the right outcome show in the above figure — for each prefix on the record with the longest sequence is retained. To implement this strategy, the FASTQ file now have to be processed twice. The first time, the records are read one by one to construct a dictionary that maps each prefix of sequences longer than $$n$$ onto the length of the longest sequence with that prefix. The second time the records are again read one by one, and only those records are retained whose sequence length matches the length onto which the prefix of the sequence is mapped by the dictionary. In case the same prefix is found in multiple records whose sequence length matches the length onto which the prefix of the sequence is mapped by the dictionary, only the first record of those records needs to be retained. For example, the sequences of records 3 and 6 in the sample FASTQ file share the same prefix and the same sequence length (the maximal length of all records sharing that prefix), and thus only record 3 needs to be retained. The sequence of record 1 also shares the same prefix, but is sequence is shorter than those of records 3 and 6.
Assignment
In this assignment we only work with FASTQ files that contain DNA sequences. These DNA sequences are represented as strings that only contain the uppercase letters A, C, G en T. Your task:
-
Write a function retain_first that takes three arguments: i) a string containing the location of a FASTQ file whose records need to be deduplicated, ii) a string containing the location of a new FASTQ file to which the retained records must be written in the same order as they appear in the given FASTQ file, and iii) the length $$n \in \mathbb{N}_0$$ of the prefixes that must be used for deduplication. The function must implement the first deduplication strategy outlined in the introduction of this assignment.
-
Write a function find_longest that takes two arguments: i) a string containing the location of a FASTQ file and ii) an integer $$n \in \mathbb{N}_0$$. The function must return a dictionary that maps each prefix of length $$n$$ from the FASTQ file (limited to prefixes of sequences having at least length $$n$$) onto the maximal length of the sequences having this prefix.
-
Use the function find_longest to write a function retain_longest. The function takes the same three arguments as the function retain_first, with the same meaning. However, this time the function must implement the second deduplication strategy outlined in the introduction of this assignment.
In practice, FASTQ files contain the results of next-generation sequencing experiments that produce millions of reads (short DNA sequences). Such files are simply too big to fit in computer memory. Therefore, avoid during the implementation of the deduplication functions to load the entire file content in computer memory, but only claim the memory needed to store the information of a single FASTQ record, together with a set (function retain_first) or a dictionary (function retain_longest) of prefixes that is used to decide whether or not the FASTQ records must be retained.
Example
In the following interactive session, we assume the FASTQ file sample.fq to be located in the current directory. This file has the same content as the sample file shown in the introduction of this assignment.
>>> retain_first('sample.fq', 'first.fq', 10)
>>> print(open('first.fq', 'r').read().rstrip())
@sample sequence 1
GGAAGTCCATGGAGTTATTTGCGGTAACCGGGACCGGCCT
+sample sequence 1
ba^aaabaa`]baaaaa_aab]D^^`b`aYDW]abaa`^a
@sample sequence 2
CTCCTTATAGCATCATACGCACAGCTGGTATCCCCATTGCTTATTAGGGTTCAGCCCTGTCGCTCTCCCC
+sample sequence 2
ababa``aaaababaaaabaa_``aa``]^aa_`aa_Z_aaaaa^baaaaaaaaaaaaaaaaaa`^```a
>>> retain_longest('sample.fq', 'longest.fq', 10)
>>> print(open('longest.fq', 'r').read().rstrip())
@sample sequence 2
CTCCTTATAGCATCATACGCACAGCTGGTATCCCCATTGCTTATTAGGGTTCAGCCCTGTCGCTCTCCCC
+sample sequence 2
ababa``aaaababaaaabaa_``aa``]^aa_`aa_Z_aaaaa^baaaaaaaaaaaaaaaaaa`^```a
@sample sequence 3
GGAAGTCCATGGAGTTATTTGCGGTAACCGGGACCGGCCTGGGGTTAGATTTTATGTCGT
+sample sequence 3
ba^aaabaa`]baaaaa_aab]D^^`b`aYDW]abaa`^ababbaaabaaaaa`]`ba`]