Point mutations occurring in DNA can be divided into two types: transitions and transversions. A transition substitutes one purine for another ($$\textrm{A} \leftrightarrow \textrm{G}$$) or one pyrimidine for another ($$\textrm{C} \leftrightarrow \textrm{T}$$). That is, a transition does not change the structure of the nucleobase. Conversely, a transversion is the interchange of a purine for a pyrimidine base, or vice-versa. See Figure 1. Transitions and transversions can be defined analogously for RNA mutations.
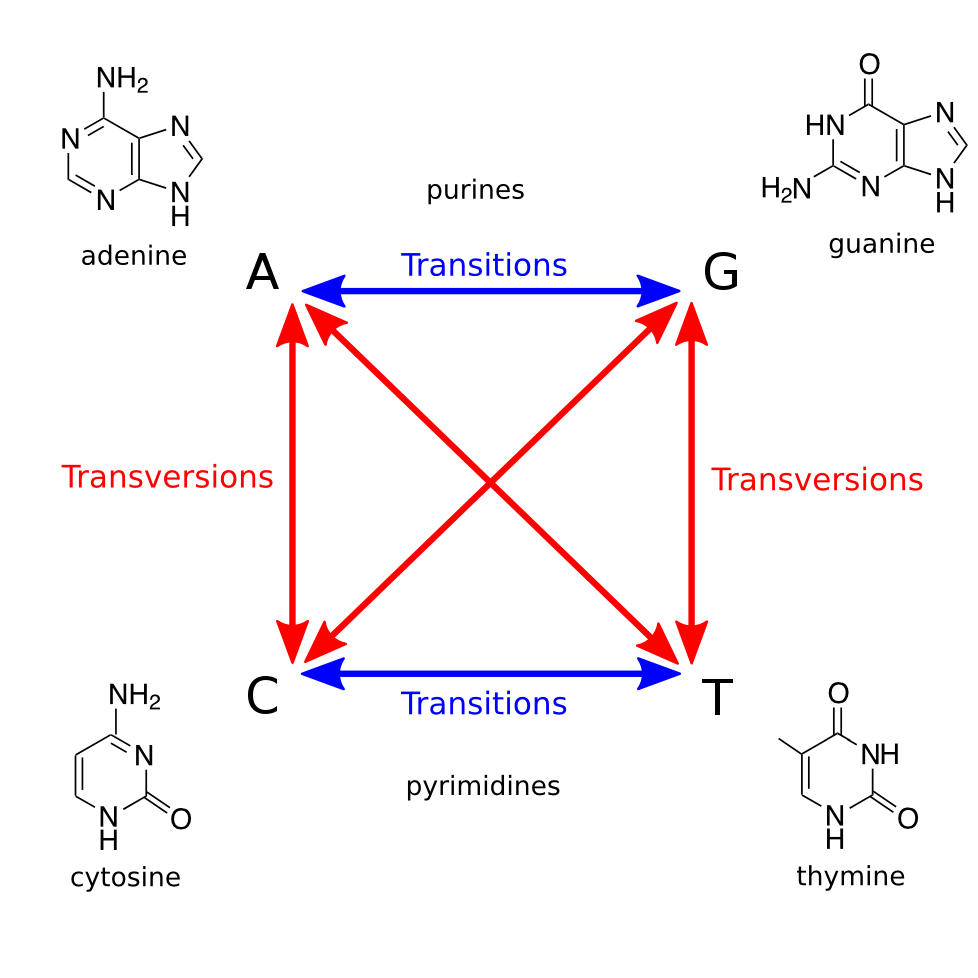
Because transversions require a more drastic change to the base's chemical structure, they are less common than transitions. Across the entire genome, the ratio of transitions to transversions is on average about 2. However, in coding regions, this ratio is typically higher (often exceeding 3) because a transition appearing in coding regions happens to be less likely to change the encoded amino acid, particularly when the substituted base is the third member of a codon (feel free to verify this fact using the DNA codon table). Such a substitution, in which the organism's protein makeup is unaffected, is known as a silent substitution.
Because of its potential for identifying coding DNA, the ratio of transitions to transversions between two strands of DNA offers a quick and useful statistic for analyzing genomes.
Assignment
For DNA strings $$s_1$$ and $$s_2$$ having the same length, their transition/transversion ratio $$R(s_1, s_2)$$ is the ratio of the total number of transitions to the total number of transversions, where symbol substitutions are inferred from mismatched corresponding symbols as when calculating Hamming distance. Your task:
-
Write a function isTransition that takes two bases $$b_1$$ and $$b_2$$. The function must return a Boolean value that indicates whether or not a mutation between $$b_1$$ and $$b_2$$ is a transition.
-
Write a function isTransversion that takes two bases $$b_1$$ and $$b_2$$. The function must return a Boolean value that indicates whether or not a mutation between $$b_1$$ and $$b_2$$ is a transversion.
-
Write a function ttRation that takes two DNA strings $$s_1$$ and $$s_2$$ of equal length. The function must return the transition/transversion $$R(s_1, s_2)$$. If the strings $$s_1$$ and $$s_2$$ do not have equal length, the function must raise an AssertionError with message strings must have equal length.
Example
In the following interactive session, we assume the FASTA file data.fna to be located in the current directory.
>>> isTransition('A', 'G') True >>> isTransition('T', 'T') False >>> isTransition('G', 'C') False >>> isTransversion('A', 'G') False >>> isTransversion('T', 'T') False >>> isTransversion('G', 'C') True >>> ttRatio('GAGCCTACTAACGGGAT', 'CATCGTAATGACGGCCT') 0.16666666666666666 >>> from Bio import SeqIO >>> ttRatio(*SeqIO.parse('data.fna', 'fasta')) 1.2142857142857142