For centuries, people have been looking for secret messages that are hidden in books, songs played backward, funny-looking Martian mensas1, or some other objects. Some believe that hidden messages in the Bible cannot be just coincidence — they must have been put in there deliberately by God himself.
The hunt for hidden messages has been popularized in modern times by the book The Bible Code2. It was written by American journalist Michael Drosnin, who claims that the Hebrew Bible contains a very complex code that reveals events that took place thousands of years after the Bible was written. Drosnin contends that some foretold events later happened exactly as predicted. Drosnin asserts that the Bible also contains hidden messages that predict the future.


To find hidden messages, Drosnin makes use of a simple technique: start at a given letter in a text and repeatedly step a fixed number of letters forward of backward. Consider the verse in the Book of Genesis (King James Version): 31:28 And hast not suffered me to kiss my sons and my daughters? Thou hast now done foolishly in so doing. If you start at the R in daughters, and skip over three letters to the O in thou, and three more to the S in hast, and so on, the hidden message Roswell is revealed! A companion hidden message — UFO — is found by starting at the U in thou, and repeatedly stepping forward 12 letters. This can't be a coincidence, no?
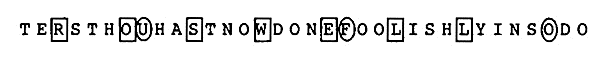
The claim that no human could have encoded the Bible in this way and that the messages cannot be just coincidence, has been questioned many times. Some critics of Drosnin say the journalist is just data mining: in any text a large amount of hidden messages can be found using Drosnin's technique and a little bit of creativity. Michael Drosnin responded to the criticism in Newsweek, by stating that "When my critics find a message about the assassination of a prime minister encrypted in Moby Dick, I'll believe them". Mathematician Brendan McKay of Australian National University and his colleagues took up the challenge, and proved that this really is not all that challenging3.
Assignment
Proteins are large biological molecules consisting of a long chain of amino acid residues. There are 20 amino acids that are used by living cells to build proteins, which are all represented by a capital (only the capitals B, J, O, U, X and Z do not correspond to an amino acid). Because the proteins that are encoded in the human genome can be represented in this was as strings of uppercase letters, Drosnin's technique is also applicable to search for hidden messages encoded in man itself.
The Latin phrase alea iacta est (the die is cast) is mainly known because it was used in 49 BC by Julius Caesar as he led his army across the river Rubicon. With this step, he entered Italy at the head of his army in defiance of the Senate and began his long civil war against Pompey and the Optimates. All words of this phrase are for example hidden in the protein sequence
HGLAVPFRTTHPSLECGRTSWARWSLDIAEFWLAWEASDCITDEDTKFQGDAVVAQM
which is part of a protein complex that allows us to smell. If we start at position 21 and successively skip 4 positions forward, we read the word ALEA. The same word can also be found by starting at the same position and successively skipping 11 positions forward. We may also start at position 36, and successively skip 11 positions backward to find a third occurrence of the word ALEA. The following table shows that the same protein also has two occurrences of the word IACTA and two occurrences of the word EST.
| start | step | length | word |
| 0 | 1 | 57 | HGLAVPFRTTHPSLECGRTSWARWSLDIAEFWLAWEASDCITDEDTKFQGDAVVAQM |
| 21 | 4 | 4 | A L E A |
| 21 | 11 | 4 | A L E A |
| 36 | -11 | 4 | A E L A |
| 27 | -6 | 5 | A T C A I |
| 27 | 6 | 5 | I A C T A |
| 29 | -10 | 3 | T S E |
| 29 | 8 | 3 | E S T |
To find similar secret messages in a given protein sequence, you may proceed as follows:
-
Write a function isaminoword that takes a string (str). The function must return a Boolean value (bool) that indicates whether the given string is an aminoword. A string is an aminoword if it has at least length two and only contains letters that are used as shorthand notation for an amino acid (all letters except B, J, O, U, X and Z). In determining whether a string is an amino word, the function should not make a distinction between uppercase and lowercase letters.
-
Write a function positions that takes two strings (str): a protein sequence and a letter. The function must return a list containing all positions in the given protein sequence where the given letter is found. In searching for letters in the protein sequence, the function should not make a distinction between uppercase and lowercase letters.
-
Write a function proteincode that takes four arguments. The first argument must be a protein sequence (str). The next three arguments should be integers (int): $$p \in \mathbb{N}$$, $$s \in \mathbb{Z}$$ and $$l \in \mathbb{N}$$. The function must return the word (str) of length $$l$$ that is read from the protein sequence by starting at position $$p$$ and successively skipping $$s$$ positions forward (or backward in case $$s$$ is negative). If it is not possible to read a word of length $$s$$ from the protein following these instructions — because the starting position does not fall within the limits of the protein or because skipping letters crosses the limits of the protein — the function must return the empty string (str).
-
Use the previous three functions to write a function proteinsearch that takes two strings (str): a protein sequence and a string. In case the given string is not an amino word (according to the definition of the function isaminoword), the function must raise an AssertionError with the message invalid aminoword. Otherwise, the function must return a list containing all possible ways the given amino word can be read from the given protein sequence. A way of reading is represented as the tuple ($$p$$, $$s$$), where $$p$$ indicates the starting position (int) and $$s$$ indicates the number of positions (int) that must be skipped forward or backward. The tuples must be sorted in increasing order in the returned list. Implement the following procedure to find all ways a given amino word can be read from a given protein sequence:
-
find all positions in the protein where the first letter of the amino word is found
-
find all positions in the protein where the second letter of the amino word is found
-
each combination of a possible position of the first and the second letter also defines the number of positions that must be skipped forward or backward in order to reach the second letter when starting from the first letter
-
check for each combination if you can read the given amino word when starting from the position of the first letter and successively skipping the computed number of positions forward or backward
In searching for occurrences of the amino word in the protein sequence, the function should not make a distinction between uppercase and lowercase letters. The function proteinsearch also has an optional parameter maxstep that takes an integer (int). In case a value is passed to this parameter, it indicates the maximal number of positions that the function may skip forward or backward in searching for occurrences of the amino words.
-
Example
>>> isaminoword('ALEA')
True
>>> isaminoword('iacta')
True
>>> isaminoword('Proline')
False
>>> protein = 'HGLAVPFRTTHPSLECGRTSWARWSLDIAEFWLAWEASDCITDEDTKFQGDAVVAQM'
>>> positions(protein, 'A')
[3, 21, 28, 33, 36, 51, 54]
>>> positions(protein, 't')
[8, 9, 18, 41, 45]
>>> proteincode(protein, 21, 11, 4)
'ALEA'
>>> proteincode(protein, 27, -6, 5)
'IACTA'
>>> proteincode(protein, 29, 8, 3)
'EST'
>>> proteincode(protein, 0, 25, 6)
''
>>> protein = 'HGLAVPFRTTHPSLECGRTSWARWSLDIAEFWLAWEASDCITDEDTKFQGDAVVAQM'
>>> proteinsearch(protein, 'ALEA')
[(21, 4), (21, 11), (36, -11)]
>>> proteinsearch(protein, 'iacta')
[(27, -6), (27, 6)]
>>> proteinsearch(protein, 'EST')
[(29, -10), (29, 8)]
>>> proteinsearch(protein, 'EST', maxstep=8)
[(29, 8)]
>>> proteinsearch(protein, 'Proline')
Traceback (most recent call last):
AssertionError: invalid amino word
Traceback (most recent call last):
AssertionError: invalid amino word