Once the complete genome of an organism has been determined, the question arises what regions of the genome are coding for proteins. To predict the exact location of these coding regions, bioinformatics tools try to infer proteins from the six reading frames. In this, a reading frame is nothing but a way to divide the sequence of nucleotides in a DNA molecule into a set of consecutive, non-overlapping triplets — in this context such a sequence of three nucleotides is called a codon.
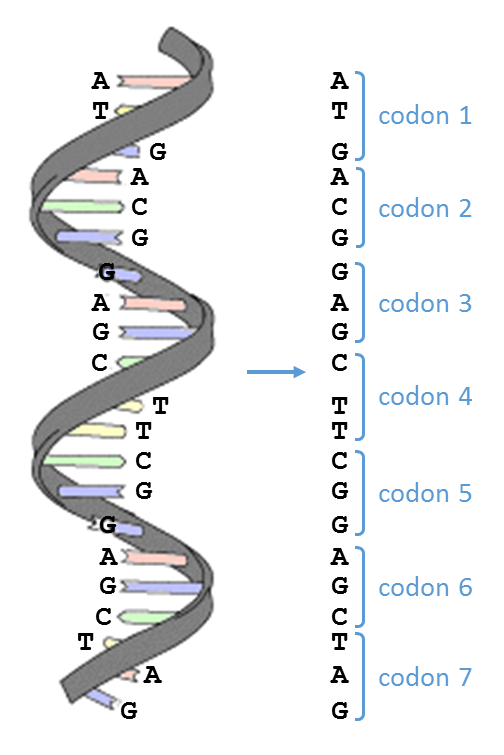
Certain codons play a special role in translating DNA into proteins:
-
start codon (ATG): the translation of a DNA sequence into a protein can start here
-
stop codon (TAG, TGA, TAA): the translation of a DNA sequence into a protein ends here
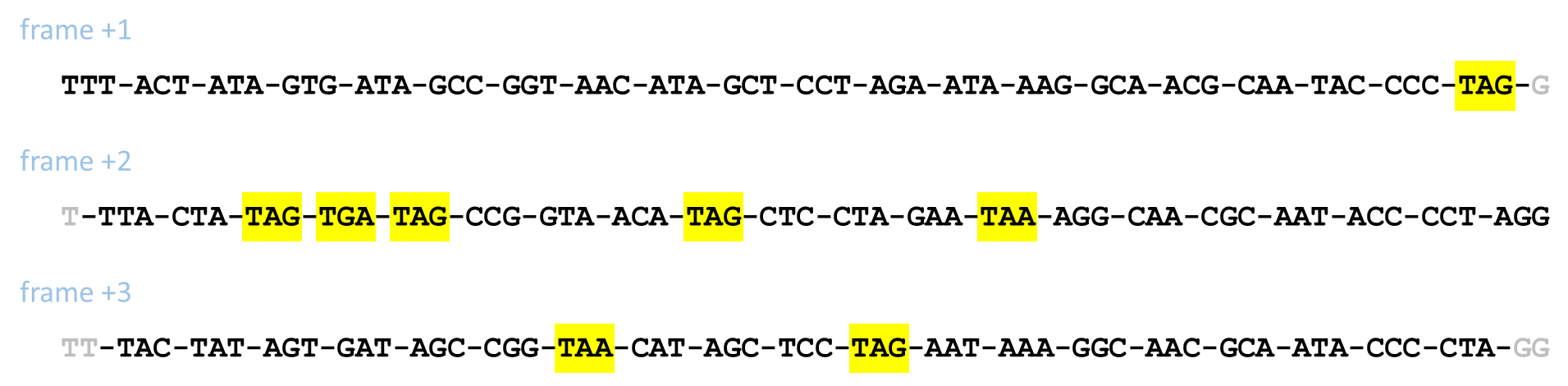
Now, the problem arises that in converting DNA into protein, translation can start at any possible position in the DNA sequence. The first codon may thus take up the first three nucleotides in the DNA sequence, but we might also skip one or two nucleotides and then the first codon takes up the next three nucleotides. As a consequence, DNA sequences can be split into codons in three possible ways, called frame +1, frame +2 and frame +3. As an example, consider the following DNA sequence
TTTACTATAGTGATAGCCGGTAACATAGCTCCTAGAATAAAGGCAACGCAATACCCCTAGG
This sequence can be split into codons in the following three ways, where
stop codons have been highlighted in yellow.
However, either strand of a DNA double helix can serve as the coding strand for proteins. Luckily, it is easy to derive the second strand from any given DNA sequence as its reverse complement. The reverse complement of a DNA string is obtained by reversing the string and taking the complement of each base symbol (A and T are complementary base symbols, as are C and G). We must reverse the string in addition to taking complements because of the directionality of DNA. DNA replication and transcription occur from the 5' end to the 3' end, and the 3' end of one strand is opposite from the 5' end of the complementary strand. Thus, if we were to simply take complements, then we would be reading the second strand in the wrong direction.
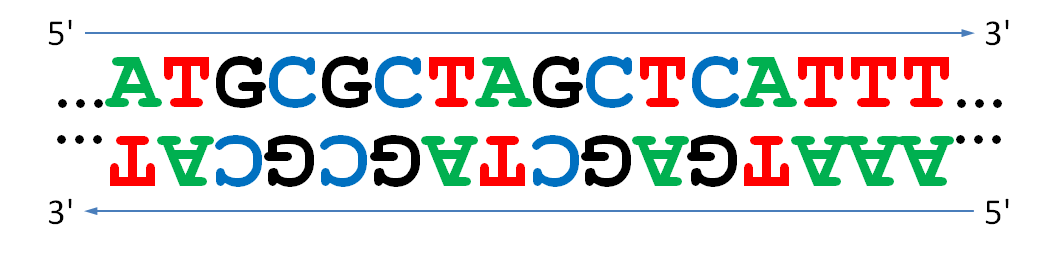
Hence, a given DNA sequence does not imply three but six reading frames in total: three reading frames result from reading the DNA sequence itself, whereas three more result from reading its reverse complement. The reverse complement is split into codons in exactly the same way as the original DNA sequence, but now these subdivisions are respectively called frame -1, frame -2 and frame -3. The reverse complement of the sample sequence is
CCTAGGGGTATTGCGTTGCCTTTATTCTAGGAGCTATGTTACCGGCTATCACTATAGTAAA
and can be split in three more frames in the following way, where stop codons have again been highlighted in yellow.

Because a reading frame that codes for a protein can not contain stop codons, detecting stop codons in the six reading frames is an important first step in determining which genomic regions code for proteins. Because no stop codons occur in reading frame -2 of the sample sequence, this is presumably the reading frame that can be used for protein translation.
Assignment
Determine the six reading frames of a given DNA sequence and count the number of stop codons in each of these reading frames. We represent DNA sequences as strings that only contain the letters A, C, G and T (both uppercase and lowercase). Your task:
-
Write a function isStopCodon that takes a DNA sequence as its argument. The function must return a Boolean value that indicates whether the given DNA sequence is a stop codon.
-
Write a function reverseComplement that takes a DNA sequence as its argument. The function must return the reverse complement of the given DNA sequence, expressed in uppercase letters.
-
Write a function stopCodons that takes two arguments: a DNA sequence and the number of a reading frame (+1, +2, +3, -1, -2 or -3). The function must return the number of stop codons that occur in the given reading frame of the given DNA sequence.
-
Write a function codons that takes two arguments: a DNA sequence and the number of a reading frame (+1, +2, +3, -1, -2 or -3). The function must return a string representation of splitting the given DNA sequence into codons in the given reading frame. This is done by separating the codons and the fragments of one or two nucleotides at the start and end of the sequence using dashes (-).
Note: In Python you can prefix any positive integer with a plus sign (called the unary plus operator1 in technical terms) to make it explicit that the number is positive. Apart from that, the numbers +42 and 42 both represent exactly the same integer value.
Example
>>> isStopCodon('TAA')
True
>>> isStopCodon('tag')
True
>>> isStopCodon('ATC')
False
>>> reverseComplement('AAGTC')
'GACTT'
>>> reverseComplement('agcttcgt')
'ACGAAGCT'
>>> reverseComplement('AGTCTTACGCTTA')
'TAAGCGTAAGACT'
>>> seq = 'TTTACTATAGTGATAGCCGGTAACATAGCTCCTAGAATAAAGGCAACGCAATACCCCTAGG'
>>> stopCodons(seq, +1)
1
>>> stopCodons(seq, +2)
5
>>> stopCodons(seq, +3)
2
>>> stopCodons(seq, -1)
3
>>> stopCodons(seq, -2)
0
>>> stopCodons(seq, -3)
1
>>> codons(seq, +1)
'TTT-ACT-ATA-GTG-ATA-GCC-GGT-AAC-ATA-GCT-CCT-AGA-ATA-AAG-GCA-ACG-CAA-TAC-CCC-TAG-G'
>>> codons(seq, +2)
'T-TTA-CTA-TAG-TGA-TAG-CCG-GTA-ACA-TAG-CTC-CTA-GAA-TAA-AGG-CAA-CGC-AAT-ACC-CCT-AGG'
>>> codons(seq, +3)
'TT-TAC-TAT-AGT-GAT-AGC-CGG-TAA-CAT-AGC-TCC-TAG-AAT-AAA-GGC-AAC-GCA-ATA-CCC-CTA-GG'
>>> codons(seq, -1)
'CCT-AGG-GGT-ATT-GCG-TTG-CCT-TTA-TTC-TAG-GAG-CTA-TGT-TAC-CGG-CTA-TCA-CTA-TAG-TAA-A'
>>> codons(seq, -2)
'C-CTA-GGG-GTA-TTG-CGT-TGC-CTT-TAT-TCT-AGG-AGC-TAT-GTT-ACC-GGC-TAT-CAC-TAT-AGT-AAA'
>>> codons(seq, -3)
'CC-TAG-GGG-TAT-TGC-GTT-GCC-TTT-ATT-CTA-GGA-GCT-ATG-TTA-CCG-GCT-ATC-ACT-ATA-GTA-AA'