Sanger sequencing is a method used to determine the DNA sequence of an organism. Developed by Frederick Sanger and colleagues in 1977, it was the most widely used sequencing method for approximately 25 years. The method is applied by radioactively or fluorescently labelling the four DNA-bases (represented by the letters A, C, G and T), so that successive bases can be read out sequentially. The radioactive or fluorescent labels can be read automatically using 4 different detectors that determine for each position in the DNA whether a radioactive or fluorescent signal is present for each of the four bases. This results in four separate signals that are then merged into a single signal, from which the DNA sequence itself may be determined.
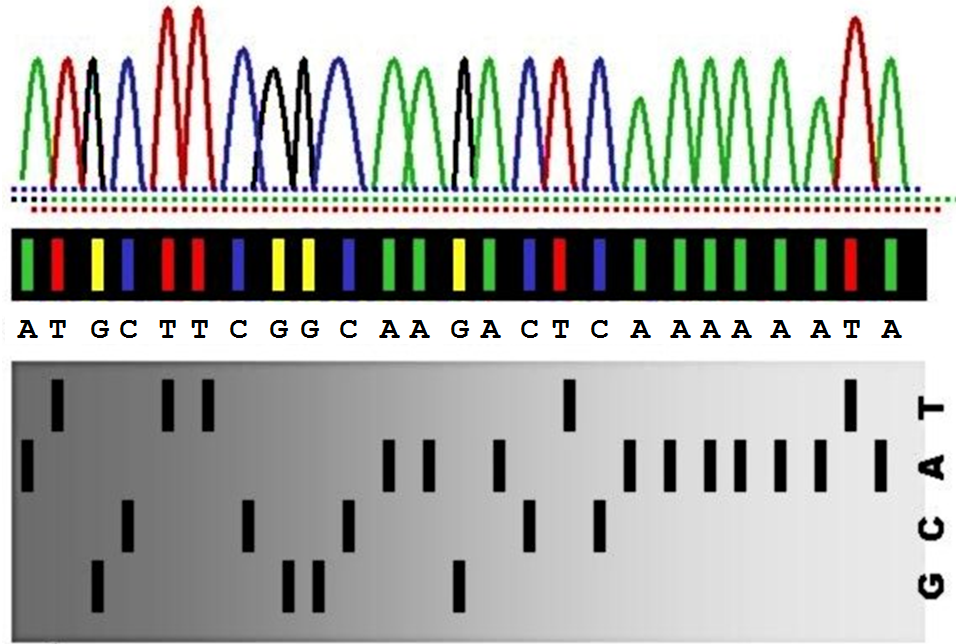
If the detector for base X (with X being one of the letters A, C, G or T) registers a positive signal at a certain position, the position is marked by the letter X. If no signal is detected, the position is marked by a hyphen. As such, the output of all four detectors — as measured for the DNA sequence ATGCTTCGGCAAGACTCAAAAAATA — is represented in the following format:
1111111111222222
1234567890123456789012345
detector A: A---------AA-A---AAAAAA-A
detector C: ---C--C--C----C-C--------
detector G: --G----GG---G------------
detector T: -T--TT---------T-------T-
=========================
DNA sequence: ATGCTTCGGCAAGACTCAAAAAATA
Assignment
Write a function sanger that takes a string argument that only contains the upper case letters A, C, G and T. This argument represents a DNA sequence. The function must return a string that represents the given DNA-sequence using the following format:
In the centre are 4 lines representing the individual signals as measures by the detector for each of the four bases A, C, G and T. These lines only contain hyphens and the base letter at the positions where a signal was measured by the detector.
Below these 4 lines is a single line that contains the same number of equality signs (=) as there are bases in the given DNA sequence.
The given DNA-sequence appears at the bottom of the format.
On top of the lines containing the individual signals are a number of lines that index the bases in the DNA-sequence. Indexing of the positions starts at 1. First there is a line containing the units of the indexes. On top of this line there is a line containing the tens, again topped by a (third) line containing the hundreds, and so on. If the length of the sequence is smaller than ten, no tens need to be represented and thus only a single line containing index digits must be included (analogous for sequences shorter than 100, 1000, …). Any position that has no digit for the tens (leading zeros are ignored) is filled with a space on the line representing the tens of the position indexes (analogous for the hundreds, thousands, ...).
Make sure that the last line returned by the function sanger, does not end with a newline. Take a look at the interactive Pyton session below to see some examples on how the formatting of the DNA-sequences must be done.
Example
>>> print(sanger('ATGCTTCGG'))
123456789
A--------
---C--C--
--G----GG
-T--TT---
=========
ATGCTTCGG
>>> print(sanger('ATGCTTCGGCAAGACTCAAAAAATA'))
1111111111222222
1234567890123456789012345
A---------AA-A---AAAAAA-A
---C--C--C----C-C--------
--G----GG---G------------
-T--TT---------T-------T-
=========================
ATGCTTCGGCAAGACTCAAAAAATA