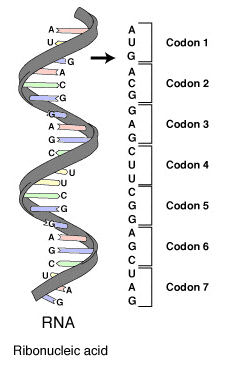
The genetic code is a set of rules indicating information encoded within genetic material (DNA or mRNA sequences) is translated into proteins (amino acid sequences) by living cells. The code defines how sequences of these nucleotide triplets (called codons) specify which amino acid will be added next during protein synthesis. For the translation of RNA into proteins we can discriminate three types of codons: translation starts with a chain initiation codon or start codon (AUG) and continues until one of the stop codons (UAG (amber), UGA (opal) or UAA (ochre)) is reached. In between start and stop codons are an arbitrary number of ordinary codons, as illustrated in the figure below.
Input
A single line containing some text that represents an RNA codon (a string that only contains the letters A, C, G and U).
Output
A single line containing the description of the type of codon read from the input. The four possible types of codons and their descriptions are given in the table below.
| type | description |
|---|---|
| start codon (AUG) | start |
| stop codon (UAG, UGA, UAA) | stop |
| ordinary codon (any other triplet of nucleotides) | normal |
| invalid codon (string of length not equal to three) | non-valid |
The output should be formatted using the following template: "The codon GCC is a normal codon.". The fragments of the template displayed in an italic font are variable, and must be filled in using the given input and the description of the codon type as determined by your program. Try to keep the number of conditions and the expression of the conditions that need to be tested as small as possible. There is no need to test whether all characters in the input string are valid bases in the RNA alphabet (A, C, G and U) in order to decide whether the codon is valid (normal, start or stop) or not.
Example
Input:
GCCOutput:
The codon GCC is a normal codon.