The genome of most bacteria consists of a single circular molecule of DNA. DNA replication — the process of producing two identical replicas from one original DNA molecule — is initiated at a particular sequence in the genome called the origin of replication, and proceeds from this point simultaneous in both directions (see figure below). The replication process ends at a position in the genome called the terminus of replication.
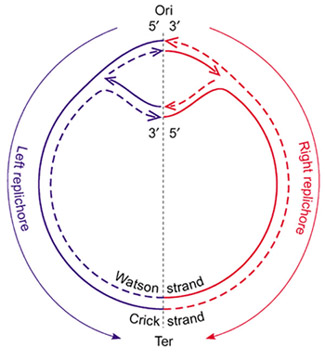
The specific structure of the origin of replication varies somewhat from species to species, but all share some common characteristics such as high AT content (adenine and thymine are easier to separate because they form only two hydrogen bonds whereas guanine and cytosine form three). The origin of replication binds the pre-replication complex, a protein complex that recognizes, unwinds, and begins to copy DNA.
A simple procedure to determine the position of the origin and terminus of replication in a genome sequence, makes use of a vector representation of the DNA molecule. This vector is constructed as a list (list) of $$(x, y)$$-coordinates (tuple; $$x, y \in \mathbb{N}$$ (int)) that starts at the point $$(0, 0)$$ in the origin. For each successive base in the DNA sequence a neighboring point is visited: the left neighbor for base A, the right neighbor for base T, the upstairs neighbor for base C and the downstairs neighbor for base G. The neighboring points are always at distance one from the previous point. The figure below shows a graphical display of the vector representation of the DNA sequence GACCCTTGT.
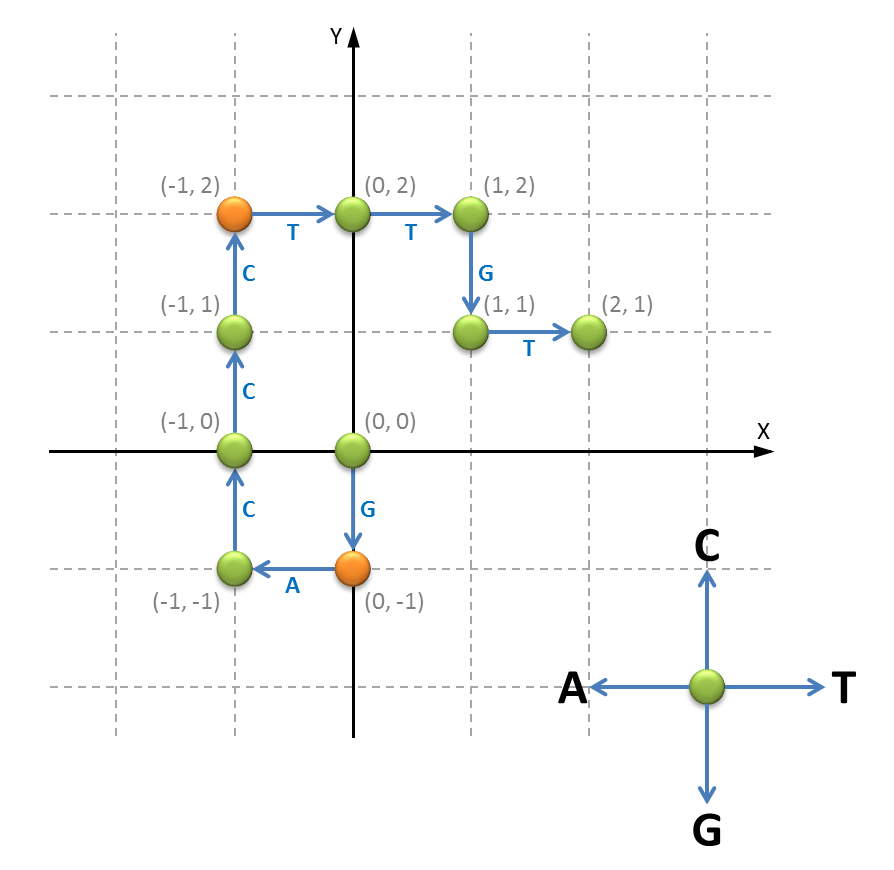
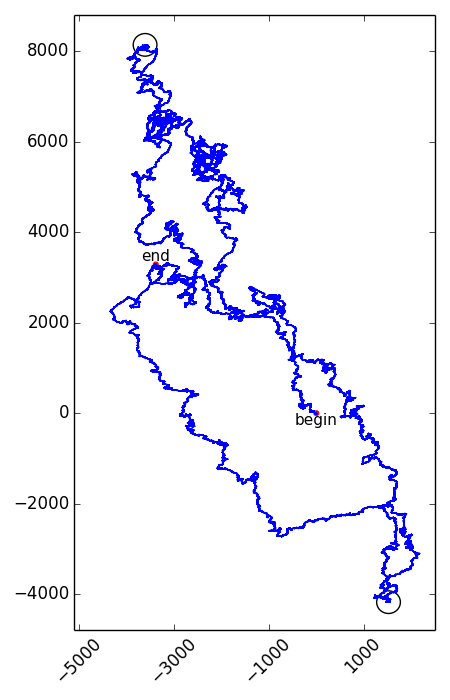
The positions in the vector representation where the $$y$$-coordinate for the first time reaches its maximal and minimal value, correspond to the positions of the origin and terminus of replication on the genome. However, which of the two points is the origin and which one is the terminus can not be determined unambiguously. Below you see an example of the vector representation of the complete genome sequence of Haemophilus influenzae strain Rd (L420231), where the positions of the origin and terminus are indicated using black circles.
Assignment
We represent DNA sequences as strings (str) that only consists of the uppercase letters A, C, G and T. Your task:
-
Write a function vector that takes a DNA sequence (str) and returns its vector representation.
-
Write a function replication that takes a DNA sequence (str). The function must return a tuple (tuple) containing two integers (int) indicating the positions in the vector representation of the DNA sequence where the $$y$$-coordinate for the first time reaches its maximal — resp. minimal — value. The positions in the vector representation are increasingly indexed starting from zero.
-
Write a function sequence that takes the vector representation of a DNA sequence and returns its corresponding DNA sequence (str).
Example
>>> vector('GACCCTTGT')
[(0, 0), (0, -1), (-1, -1), (-1, 0), (-1, 1), (-1, 2), (0, 2), (1, 2), (1, 1), (2, 1)]
>>> vector('CTGGGGTAA')
[(0, 0), (0, 1), (1, 1), (1, 0), (1, -1), (1, -2), (1, -3), (2, -3), (1, -3), (0, -3)]
>>> replicatie('GACCCTTGT')
(5, 1)
>>> replicatie('CTGGGGTAA')
(1, 6)
>>> sequentie([(0, 0), (0, -1), (-1, -1), (-1, 0), (-1, 1), (-1, 2), (0, 2), (1, 2), (1, 1), (2, 1)])
'GACCCTTGT'
>>> sequentie([(0, 0), (0, 1), (1, 1), (1, 0), (1, -1), (1, -2), (1, -3), (2, -3), (1, -3), (0, -3)])
'CTGGGGTAA'Resources
-
Lobry JR (1996). A simple vectorial representation of DNA sequences for the detection of replication origins in bacteria. Biochimie 78, 323-326. 3
-
Mackiewicz P, Mackiewicz D, Kowalczuk M, Cebrat S (2001). Flip-flop around the origin and terminus of replication in prokaryotic genomes. Genome Biology 2(12). 4