A notorious problem in molecular biology,is protein folding. A protein molecule consists of a chain of amino acids that spontaneously folds into a three-dimensional shape that is more energetically favorable . This binding energy depends on the mutual position of all amino acids. This provides so many options that even the most advanced supercomputers still crash here.
In order to make the problem somewhat easier we are going to simulate a protein molecule that folds in two dimensions. The initial state is a square grid in which each amino acid has a positive or negative charge. The folding is done by placing baffles between the boxes in such a way that a circuit is created through the grid that includes all the booths. The ends of the molecule do not have to be situated at the edge of the grid.
Whenever a baffle is positioned between two boxes, the molecule is folded and the amino acids are touching each other. If there is a baffle between a box with charge $$-3$$ and a box with charge $$4$$, this results in a binding energy of $$-3 \times 4 = -12$$. An amino acid can affect several other amino acids, and then yield binding energy with every neighbour. Of course, due to the minus signs, binding energy can be both positive and negative.
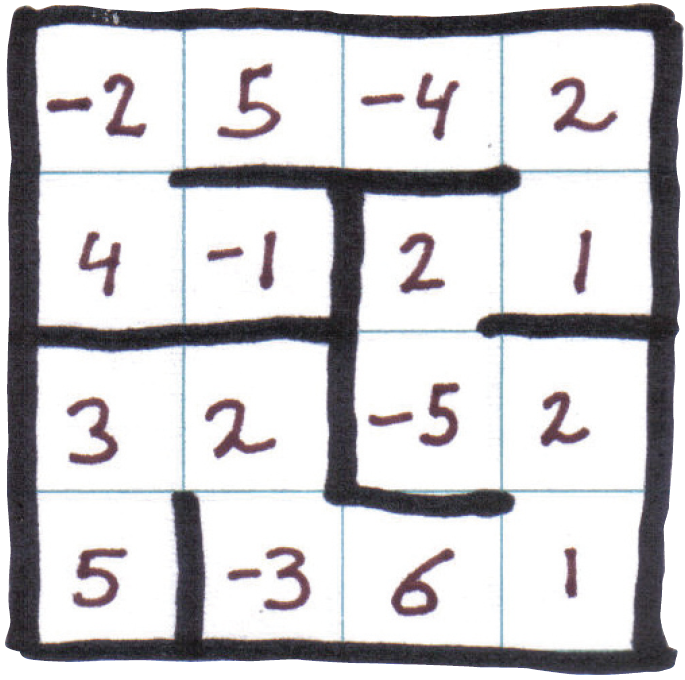
For example, you can see a $$4\times4$$ grid above, in which a protein with 16 amino acids is folded to a configuration with binding energy equal to \[ \begin{multline} \phantom{xxxxxxxxxxxx}(-1 \times 2) + (2 \times -5) + (5 \times -3) + (4 \times 3) + (5\times -1) + \phantom{x}\\ (-1 \times 2) + (-4 \times 2) + (-5 \times 6) + (1 \times 2) = -58\phantom{xxxxxxxxxxxx} \nonumber \end{multline} \]
Assignment
Define a class Protein with the following methods:
An initializing method __init__ which can be used to make a protein with a given initial state. This initial stated must be given to the initializing method as a first, obligatory parameter, in the format of a list of whole numbers. The initializing method must verify whether a square grid can be made from this list. If this is not the case, an AssertionError must occur as indicated in the example below. Otherwise, the initial state must be saved in a the newly made Protein object.
A method fold with an optional parameter with which a fold configuration for the protein can be given in the format of a list of tuples. In that list, every tuple has the format ($$r_1, k_1, r_2, k_2$$) and indicates that a baffle must be placed between two neighbouring box $$(r_1, k_1)$$ and $$(r_2, k_2)$$ when folding. Here, $$r$$ and $$k$$ respectively represent a row number and a column number, where the numbering always starts from aero. The method may assume that a list of tuples is given as fold configuration, and that every tuple consists of four whole numbers. The method does have to verify for every tuple if these four numbers represent two valid positions of boxes in the square grid, and that the first box is situated either on the left of the second box in the grid, or directly above. Look at the example below to see how the method must react if an invalid fold configuration was given. If a valid value was given to this optional parameter, this configuration must be kept by the Protein object. A possible former configuration is then overwritten by the new configuration. If no value was given to the optional parameter, the protein is not folded (in other words, there are no boxes).
In order to immediately built a Protein object with a given initial state and an initial folding configuration, a folding configuration can be given to the initializing method __init__ as a second optional parameter. This has the same format and meaning of the parameter from the method fold. If a value is given for this parameter, the folding configuration must be set for the protein by calling the method fold. Otherwise, no folding configuration is initially set for the protein.
A method bindingenergy that prints the binding energy of the protein. This binding energy must be calculated based on the folding configuration that was set for the protein. If no folding configuration was set, 0 (zero) must be printed.
A method __str__ which can be used to print the string representation of the protein of the format of the example below. Here, the charges of the amino acids must be shown centrated over four positions in the boxes. If a folding configuration was set, the baffles must be shown in the right place.
Example
>>> protein = Protein([-2, 5, -4, 2, 4, -1, 2])
Traceback (most recent call last):
AssertionError: invalid initial state
>>> protein = Protein([-2, 5, -4, 2, 4, -1, 2, 1, 3, 2, -5, 2, 5, -3, 6, 1])
>>> eiwit.bindingenergy()
0
>>> print(protein)
+----+----+----+----+
| -2 5 -4 2 |
+ + + + +
| 4 -1 2 1 |
+ + + + +
| 3 2 -5 2 |
+ + + + +
| 5 -3 6 1 |
+----+----+----+----+
>>> protein.fold([(1, 1, 1, 2), (2, 1, 2, 2), (3, 0, 3, 1),
... (1, 0, 2, 0), (0, 1, 1, 1), (1, 1, 2, 1),
... (0, 2, 1, 2), (2, 2, 3, 2), (1, 3, 2, 3)])
>>> protein.bindingenergy()
-58
>>> print(protein)
+----+----+----+----+
| -2 5 -4 2 |
+ +----+----+ +
| 4 -1 | 2 1 |
+----+----+ +----+
| 3 2 | -5 2 |
+ + +----+ +
| 5 | -3 6 1 |
+----+----+----+----+
>>> protein.fold([(1, 1, 1, 2), (2, 1, 2, 2), (4, 0, 4, 1)])
Traceback (most recent call last):
AssertionError: invalid configuration
>>> protein.fold([(1, 1, 1, 2), (2, 1, 3, 3), (3, 0, 3, 1)])
Traceback (most recent call last):
AssertionError: invalid configuration
>>> protein = Protein([7, -2, -4, 3], [(0, 1, 1, 1)])
>>> protein.bindingenergy()
-6
>>> print(protein)
+----+----+
| 7 -2 |
+ +----+
| -4 3 |
+----+----+