Breaking the bonds
In "Perfect matchings and RNA secondary structures" we considered a problem that required us to assume that every possible nucleotide is involved in base pairing to induce an RNA secondary structure. Yet the only way this could occur is if the frequency of adenine in our RNA strand is equal to the frequency of uracil and if the same holds for guanine and cytosine.
We will therefore begin to explore ways of counting secondary structures
in which this condition is not required. A more general combinatorial
problem will ask instead for the total number of secondary structures of a
strand having a maximum possible number of base pairs.
Assignment
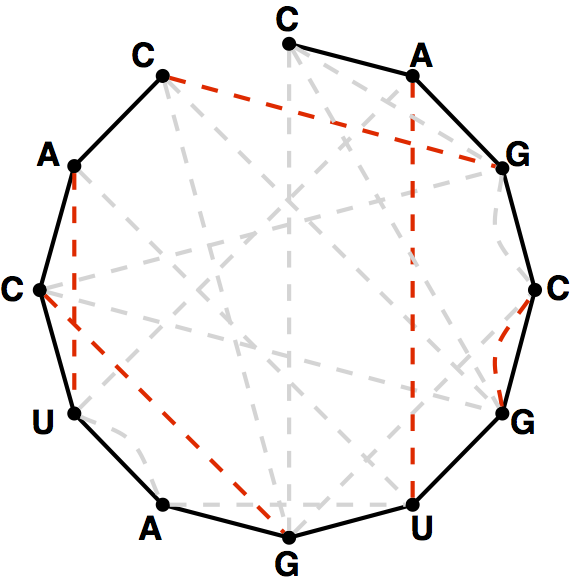
The graph theoretical analogue of the quandary stated in the introduction above is that if we have an RNA string $$s$$ that does not have the same number of occurrences of C as G and the same number of occurrences of A as U, then the bonding graph of $$s$$ cannot possibly possess a perfect matching among its basepair edges. In fact, most bonding graphs will not contain a perfect matching.
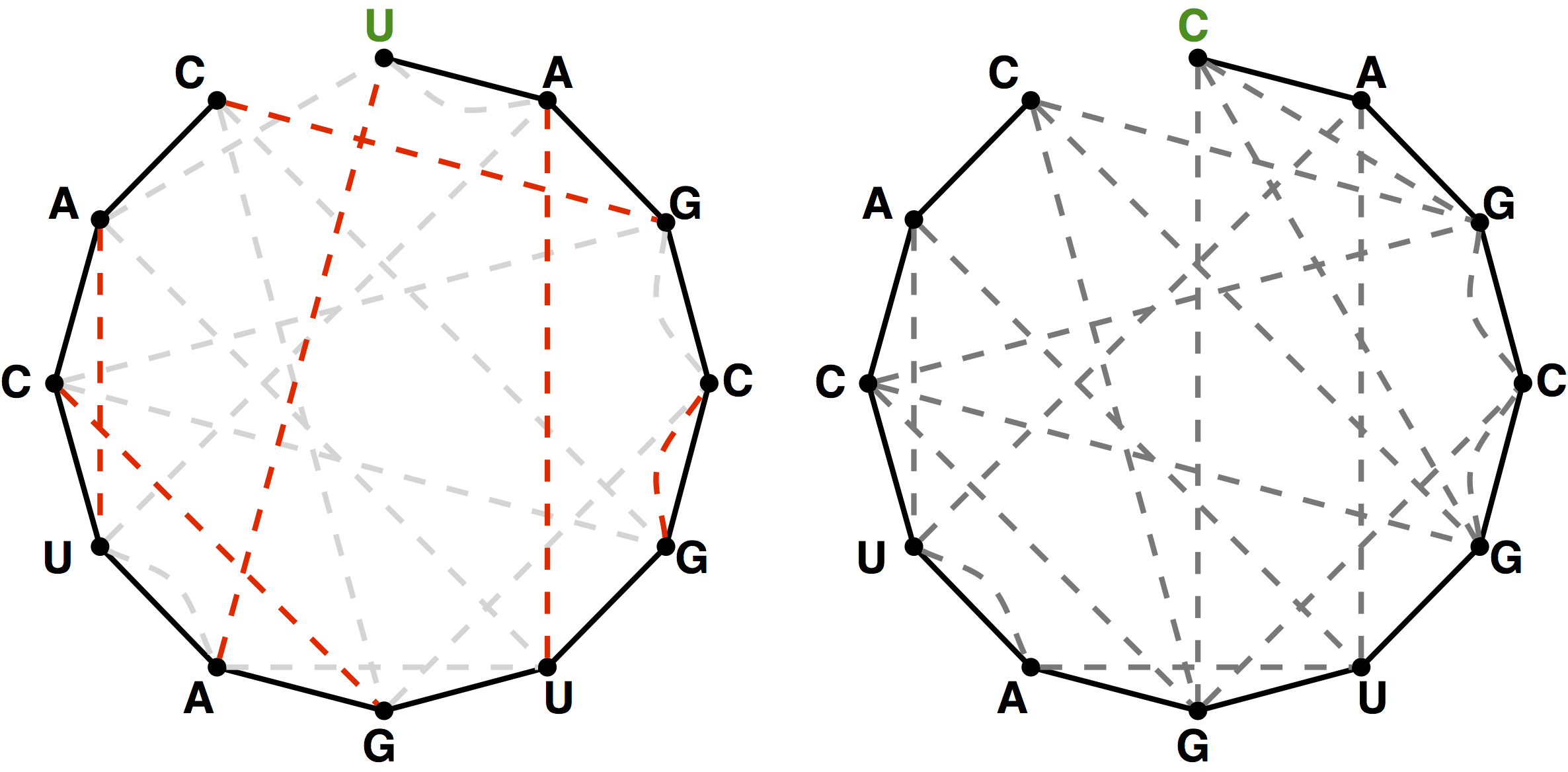
In light of this fact, we define a maximum matching in a graph as a matching containing as many edges as possible.
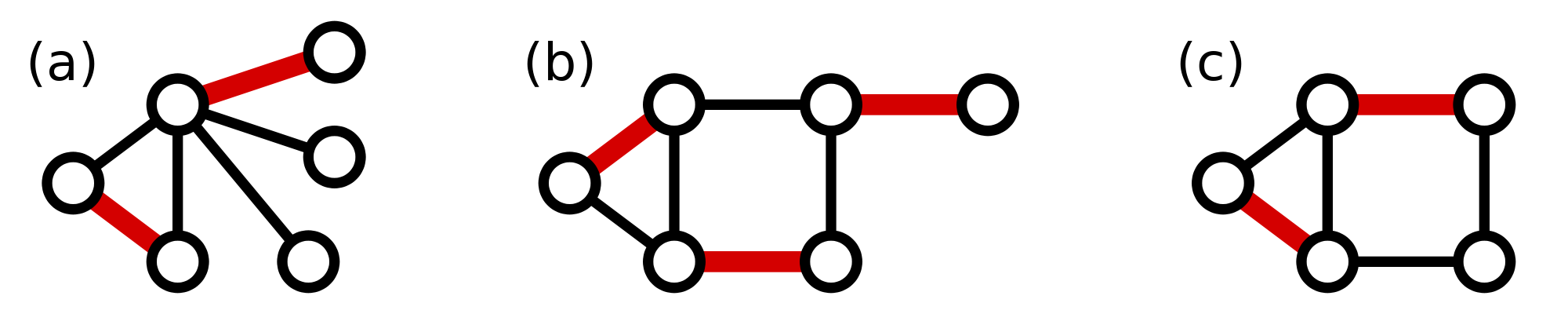
A maximum matching of basepair edges will correspond to a way of forming
as many base pairs as possible in an RNA string.
Write a function maximumMatchings that takes an RNA strings $$s$$. The function must return the total possible number of maximum matchings of basepair edges in the bonding graph of $$s$$.
Example
In the following interactive session, we assume the FASTA file data.fna to be located in the current directory.
>>> maximumMatchings('AUGCUUC') 6 >>> from Bio import SeqIO >>> maximumMatchings(*SeqIO.parse('data.fna', 'fasta')) 14237240361899797123612803072000000000