The war between viruses and bacteria has been waged for over a billion years. Viruses called bacteriophages1 (or simply phages) require a bacterial host to propagate, and so they must somehow infiltrate the bacterium. Such deception can only be achieved if the phage understands the genetic framework underlying the bacterium's cellular functions. The phage's goal is to insert DNA2 that will be replicated within the bacterium and lead to the reproduction of as many copies of the phage as possible, which sometimes also involves the bacterium's demise.
To defend itself, the bacterium must either obfuscate its cellular functions so that the phage cannot infiltrate it, or better yet, go on the counterattack by calling in the air force. Specifically, the bacterium employs aerial scouts called restriction enzymes3, which operate by cutting through viral DNA to cripple the phage. But what kind of DNA are restriction enzymes looking for?
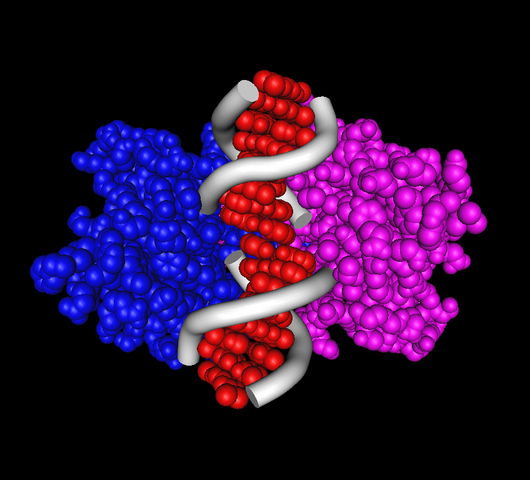
The restriction enzyme is a homodimer4, which means that it is composed of two identical substructures. Each of these structures separates from the restriction enzyme in order to bind to and cut one strand of the phage DNA molecule. Both substructures are pre-programmed with the same target string containing 4 to 12 nucleotides to search for within the phage DNA (see figure above). The chance that both strands of phage DNA will be cut (thus crippling the phage) is greater if the target is located on both strands of phage DNA, as close to each other as possible. By extension, the best chance of disarming the phage occurs when the two target copies appear directly across from each other along the phage DNA, a phenomenon that occurs precisely when the target is equal to its own reverse complement5. Eons of evolution have made sure that most restriction enzyme targets now have this form.

Assignment
In this assignment we represent a DNA sequence as a string that only contains the uppercase letters A, C, G and T. The reverse complement6 of formed by reversing the string and taking the complement of each character. The characters A and T are complement each other, and so do the characters C and G. We must also reverse the string in addition to taking complements because of the directionality of DNA: DNA replication and transcription occurs from the 3' end to the 5' end, and the 3' end of one strand is opposite from the 5' end of the complementary strand. Thus, if we were to simply take complements, then we would be reading the second strand in the wrong direction.
A DNA sequence is a reverse palindrome7 if it is equal to its reverse complement. For instance, GCATGC is a reverse palindrome because its reverse complement is GCATGC (see figure above). Your task:
-
Write a function reverseComplement that takes a DNA sequence. The function must return the reverse complement of the given DNA sequence.
-
Write a function reversePalindrome that takes a DNA sequence. The function must return a Boolean value that indicates whether or not the given DNA sequence is a reverse palindrome.
-
Write a function restrictionSites that takes a DNA sequence. The function must return a list containing all restriction sites in the given DNA sequence. A restriction site is a position in a DNA sequence where a reverse palindrome is located. Each restriction site is represented by a tuple that contains the position of the first letter of the palindrome, together with the palindrome itself. Here we assume that the first character of the DNA sequence is at position 1, the second letter at position 2, and so on. The restriction sites must be sorted, first according to increasing start position and then according to increasing length of the palindromes. The function has two additional optional arguments minLength (default value: 4) and maxLength (default value: 12) that respectively take the minimal and maximal length of the palindromes that must be taken into account to determine the restriction sites.
Example
>>> reverseComplement('GATATC')
'GATATC'
>>> reverseComplement('GCATGC')
'GCATGC'
>>> reverseComplement('AGCTTC')
'GAAGCT'
>>> reversePalindrome('GATATC')
True
>>> reversePalindrome('GCATGC')
True
>>> reversePalindrome('AGCTTC')
False
>>> restrictionSites('TCAATGCATGCGGGTCTATATGCAT')
[(4, 'ATGCAT'), (5, 'TGCA'), (6, 'GCATGC'), (7, 'CATG'), (17, 'TATA'), (18, 'ATAT'), (20, 'ATGCAT'), (21, 'TGCA')]
>>> restrictionSites('AAGTCATAGCTATCGATCAGATCAC', minLength=5)
[(6, 'ATAGCTAT'), (7, 'TAGCTA'), (12, 'ATCGAT')]
>>> restrictionSites('ATATTCAGTCATCGATCAGCTAGCA', maxLength=5)
[(1, 'ATAT'), (12, 'TCGA'), (14, 'GATC'), (18, 'AGCT'), (20, 'CTAG')]Epilogue
You may be curious how the bacterium prevents its own DNA8 from being cut by restriction enzymes. The short answer is that it locks itself from being cut through a chemical process called DNA methylation9. DNA methylation is a chemical process that a cell applies to its own DNA by bonding methyl groups ($$CH_3$$) to nucleotides10, which effectively locks them from being involved in a reaction (especially those involving further bonding, like transcription11).
Methylation serves a number of fascinating practical purposes. In one example, restriction enzymes13 employed by a bacterium would not be capable of discriminating between the foreign DNA of a phage14 and the bacterium's own DNA, so the bacterium methylates its DNA to protect it from its own restriction enzymes.
Methylation is also a remarkable way to regulate gene activity15, as methylated DNA can be inherited, which has opened up a brand new field called epigenetics16. This field studies functionally relevant modifications to the genome17 that do not involve a change in the genome's sequence of nucleotides. In short, the ultimate truth is that there is a lot more to inheritance than simply replicating DNA18!
Methylation usually occurs at CpG sites19, where cytosine20 and guanine21 nucleotides appear consecutively. In recent years, researchers have shown that DNA methylation occurs in higher organisms and that it is important for normal development: methylated areas of the genome are protected from transcription activators22 and remain inactive. These "silent" parts of the genome are called heterochromatin23.