Given a collection of DNA fragments from a large DNA sequence. For each fragment, a list of other fragments which are positioned to the right of this fragment are given. A consistent ordering is one that respects these constraints.
E.g., given the fragments (ACA, ATT, GGC, CCA, AAT) and the following constraints:
- to the right of ACA are ATT and GGC,
- to the right of ATT are GGC and CCA,
- to the right of GGC is AAT,
- to the right of CCA are GGC and AAT
The constraints can be represented by a directed acyclic graph:
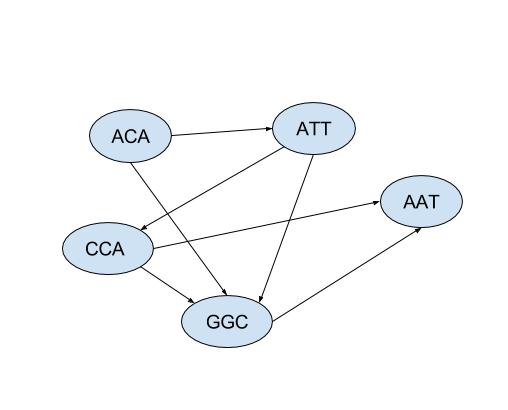
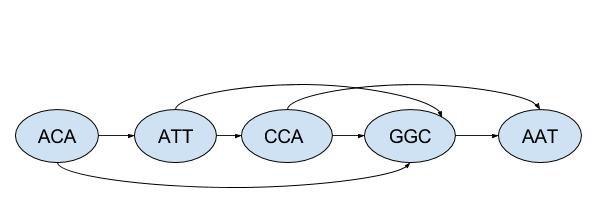
A consistent ordering is [ACA, ATT, CCA, GGC, AAT]. The corresponding drawing of the graph has all arrows pointing forward:
Assignment
Write a Python function ordering which takes a collection of DNA fragments as parameter and which returns a list with a consistent ordering of the fragments.
Example
>>> ordering({'ACA': ['ATT', 'GGC'], 'ATT':['GGC', 'CCA'], 'GGC': ['AAT'], 'CCA': ['GGC', 'AAT']})
['ACA', 'ATT', 'CCA', 'GGC', 'AAT']