Evolution as a sequence of mistakes
A mutation is simply a mistake that occurs during the creation or copying of a nucleic acid, in particular DNA. Because nucleic acids are vital to cellular functions, mutations tend to cause a ripple effect throughout the cell. Although mutations are technically mistakes, a very rare mutation may equip the cell with a beneficial attribute. In fact, the macro effects of evolution are attributable by the accumulated result of beneficial microscopic mutations over many generations.
The simplest and most common type of nucleic acid mutation is a point mutation, which replaces one base with another at a single nucleotide. In the case of DNA, a point mutation must change the complementary base accordingly.
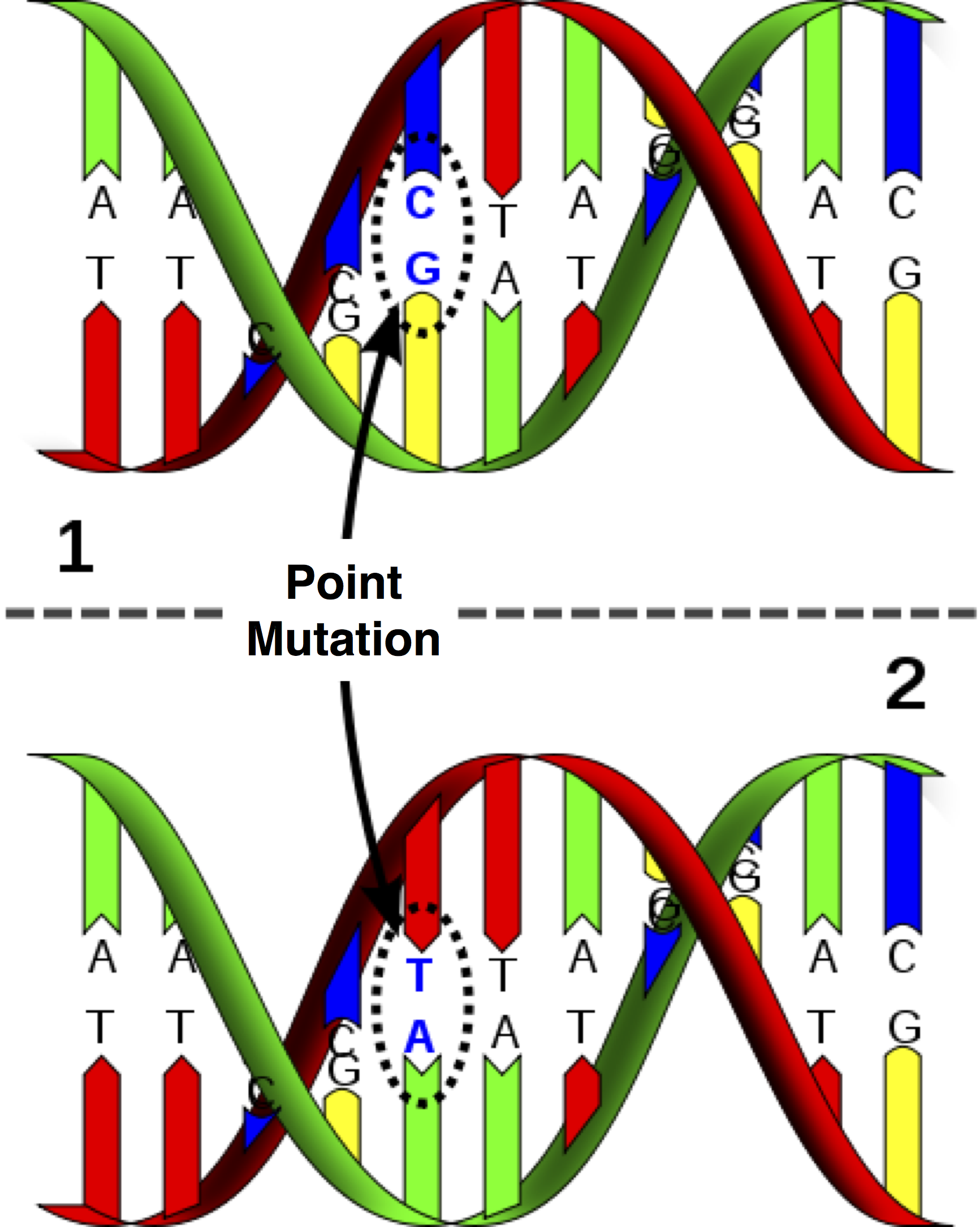
Two DNA strands taken from different organism or species genomes are homologous if they share a recent ancestor. Thus, counting the number of bases at which homologous strands differ provides us with the minimum number of point mutations that could have occurred on the evolutionary path between the two strands.
We are interested in minimizing the number of (point) mutations separating two species because of the biological principle of parsimony, which demands that evolutionary histories should be as simply explained as possible.Assignment
Given two strings $$s_1$$ and $$s_2$$ of equal length, the Hamming distance between $$s_1$$ and $$s_2$$, denoted $$d_H(s_1, s_2)$$, is the number of corresponding symbols that differ in $$s_1$$ and $$s_2$$.
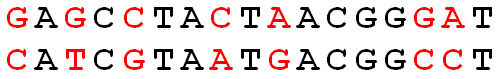
Write a function hammingDistances that takes two DNA strings $$s_1$$ and $$s_2$$ of equal length. The function must return the Hamming distance $$d_H(s_1, s_2)$$. If the strings $$s_1$$ and $$s_2$$ do not have equal length, the function must raise an AssertionError with message strings must have equal length.
Example
In the following interactive session, we assume the FASTA file data.fna to be located in the current directory.
>>> hammingDistance('GAGCCTACTAACGGGAT', 'CATCGTAATGACGGCCT') 7 >>> from Bio import SeqIO >>> hammingDistance(*SeqIO.parse('data.fna', 'fasta')) 31