Huntington's disease, which was reported thoroughly by the American doctor George Huntington in 1872, is a dominant hereditary condition which affects certain parts of the brain. The first symptoms of HD are mostly displayed between the age of 35 and 45, and, among others, consist of uncontrolled (choreatic) movements that slowly get worse, mental deterioration and a variety of psychiatric disorders. On average, the disease is fatal after eighteen years, mainly because of additional causes such as pneumonia.
HD is classified under the trinucleotide repeat disorders, which are caused by a repeated section of a gene, causing a deviation from the normal length. With HD it concerns a deviation in the Huntingtin gene. The Huntingtin gene exhibits a sequence of three base pairs at the 5' end of the DNA — cytosine-adenine-guanine (CAG) coding for the amino acid glutamine — that is repeated multiple times (…CAGCAGCAG…). This region is called a trinucleotide repeat. The figure below shows a distribution of the normal and expanded length of the HD trinucleotide repeat.
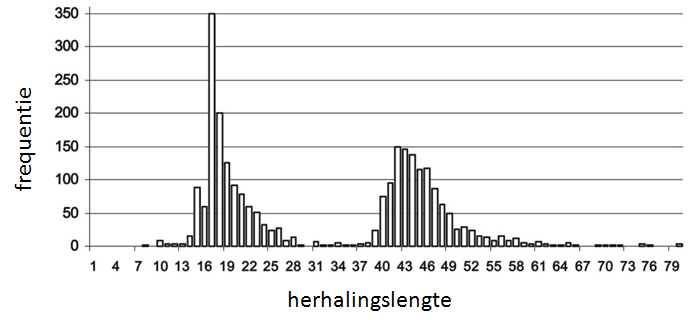
The distribution of repeats for the Huntington's disease can be divided into four categories. Repeats of 26 or less are perfectly normal. repeats between 27 and 35 are rare and are not associated with expression of the disease, but occasionally fathers with repeats will transfer a repeat to their heirs which is expanded within the interval for expression of the disease. Repeats between 36 and 39 are associated with reduced penetrance, in which case some people will develop HD and others will not. Repeats of 40 or more are associated with the expression of HD. Individuals who carry repeats in this category will develop HD, assuming they do not die earlier on in their life by other causes.
repeatlength diagnosis <27 normal 27-35 low risk 36-39 increased risk >39 absolute risk
Assignment
Write a function repeatlength, that determines the maximum sequence of repeats of a given string B for a given string A. The length of the two given strings is variable, and the comparison of both strings has to be executed without differentiating between uppercase and lowercase letters. The table below shows the number of examples of parameter values, and the matching result that should be generated by the function.
If string B does not occur in string A, then the function should return the value zero. If the string A contains several subsequences consisting of repeats of the string B, then the number of repeats of the longest subsequence should be returned. Repeats never overlap, which means, for example, the string TTTT contains two repeats of the string TT and not three.string A string B result AATCGTCGTCGTAGCTTCGTGGTGAAGATAG CTGTA 0 AATCGTCGTCGTAGCTTCGTGGTGAAGATAG gtg 2 aatcgtcgtcgtagcttcgtggtgaagatag TCG 3 Write a function HuntingtonDiagnosis, that gives a diagnosis for the given DNA sequence of a Huntington gene concerning the possible risk for the development of Huntington's disease. This diagnosis is of course dependent on the number of repeats of the trinucleotide CAG (use the function repeatlength), and should be returned as a string similar to the table of diagnosis above. A DNA sequence is represented by a string containing only the lettes A, G, C and T (both uppercase and lowercase letters are allowed).
Example
>>> repeatlength("AATCGTCGTCGTAGCTTCGTGGTGAAGATAG","CTGTA")
0
>>> repeatlength("AATCGTCGTCGTAGCTTCGTGGTGAAGATAG","gtg")
2
>>> repeatlength("aatcgtcgtcgtagcttcgtggtgaagatag","TCG")
3
>>> HuntingtonDiagnosis('CAG' * 20)
'normal'
>>> HuntingtonDiagnosis('CAG' * 35)
'low risk'
>>> HuntingtonDiagnosis('CAG' * 38)
'increased risk'
>>> HuntingtonDiagnosis('CAG' * 52)
'absolute risk'