Finding mutated motifs
We have discussed at length the importance of motif finding in biology for genetic strings. However, searching for exact substring matches is of little use in applications because a motif can vary under the effect of mutation. Fortunately, we already possess functions like edit distance for quantifying the similarity of two strings.
Furthermore, recall that each chromosome is made up of a large number of genes (on average, each human chromosome contains over 1,000 genes). Therefore, to determine whether a newly sequenced chromosome contains a given gene, neither local nor global alignment applies.
One possible alignment variant for finding genes is semiglobal alignment, which we discuss in "Semiglobal alignment1". Yet semiglobal alignment only allows us to disregard gaps at the end of the alignment. To find a known gene in a new chromosome, we need to instead align the gene against intervals of the chromosome, a problem that calls for an entirely new algorithmic variation of alignment.
Assignment
Given a string $$s$$ and a motif $$t$$, an alignment of a substring of $$s$$ against all of $$t$$ is called a fitting alignment. Our aim is to find a substring $$s'$$ of $$s$$ that maximizes an alignment score with respect to $$t$$.
Note that more than one such substring of $$s$$ may exist, depending on the particular strings and alignment score used. One candidate for scoring function is the one derived from edit distance. In this assignment, we will consider a slightly different alignment score, in which all matched symbols count as +1 and all mismatched symbols (including insertions and deletions) receive a cost of -1. Let's call this scoring function the mismatch score. See the figure below for a comparison of global, local, and fitting alignments with respect to mismatch score.
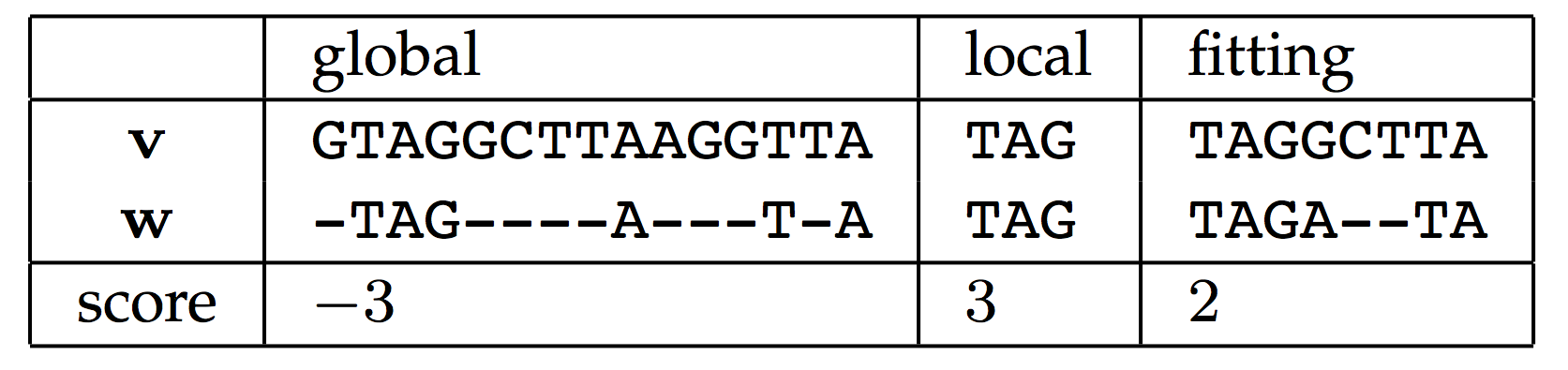
Your task:
-
Write a function fittingAlignmentScore that takes a DNA strings $$s$$ representing a genome and a DNA string $$t$$ representing a motif. The function must return the optimal fitting alignment score with respect to the mismatch score defined above.
-
Write a function fittingAlignment that takes a DNA strings $$s$$ representing a genome and a DNA string $$t$$ representing a motif. The function must return an optimal fitting alignment of a substring of $$s$$ against $$t$$ with respect to the mismatch score defined above. If multiple such alignments exist, then the function may return any one.
Example
In the following interactive session, we assume the FASTA file data.fna2 to be located in the current directory.
>>> from Bio import SeqIO >>> fittingAlignmentScore('GCAAACCATAAGCCCTACGTGCCGCCTGTTTAAACTCGCGAACTGAATCTTCTGCTTCACGGTGAAAGTACCACAATGGTATCACACCCCAAGGAAAC', 'GCCGTCAGGCTGGTGTCCG') 5 >>> fittingAlignmentScore(*SeqIO.parse('data.fna', 'fasta')) 9 >>> fittingAlignment('GCAAACCATAAGCCCTACGTGCCGCCTGTTTAAACTCGCGAACTGAATCTTCTGCTTCACGGTGAAAGTACCACAATGGTATCACACCCCAAGGAAAC', 'GCCGTCAGGCTGGTGTCCG') ('GCCCT-A--C-G-TG-CCG', 'GCCGTCAGGCTGGTGTCCG') >>> fittingAlignment(*SeqIO.parse('data.fna', 'fasta')) ('GCA-TGGGTTGTTAGGGATTGCGTTAG', 'GAAATG--TTG--AGCGAGT-CGTTAG')