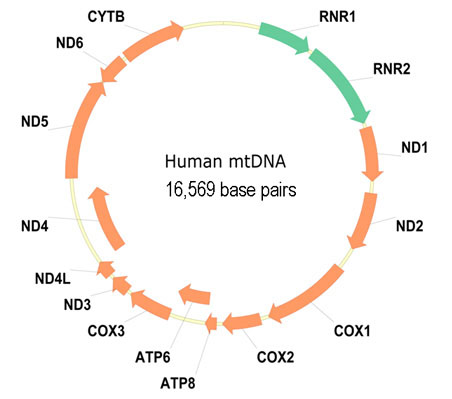
The genome of an organism is the whole of hereditary infromation in a cell. This hereditary information is either coded in DNA or — for some types of viruses — in RNA. Genes are structural components of DNA and RNA that code for a polypeptide or an RNA chain that has a certain function in an organism. The image below, for example, shows the different genes that are situated on a ring-shaped mitochondrial DNA (mrDNA) of a human being. The arrows that are used in the representation of genes, indicate that they can be oriented both forward and backward (DNA is double-stranded). A gene is localized on a genome by giving the position of the first and last base of the gene. Positions are indicated with regard to the start of the genome (for ring-shaped genomes, an initial position is chosen), that gets index 1.
What is unclear in the image above, is that genes can also (partially) overlap. This is clear in the structure of the HIV virus (an RNA lentivirus that causes AIDS) that is given below. The genes are indicated as rectangles (RNA has only one string, which results in all genes being oriented forward).
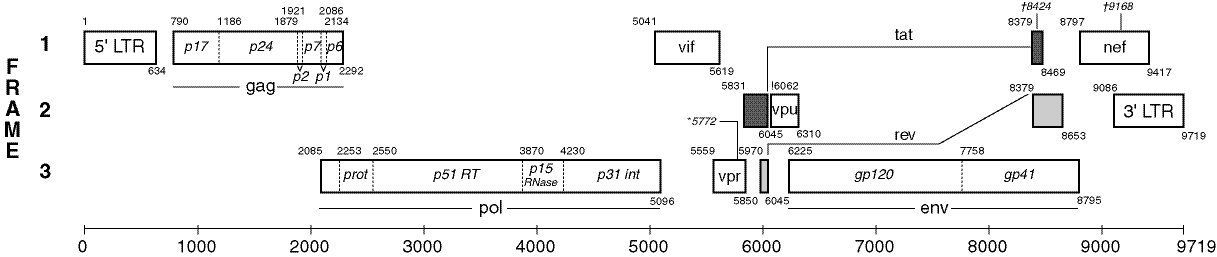
The density of a genome is a percentage that indicates how many positions within the genome are taken by genes (or other structural elements). The genome density can be determined by summing up the lengths of the genes and then dividing it by the length of the genome. This, however, is a naive way of working, that doesn't take the overlapping genes into account and consequentially calculates some positions multiple times. A better way consists of determining what the percentage of the positions within the genome that are situated in at least one gene.
Assignment
Define a class Gene which can be used to represent gene objects that contains the following methods:
An initializing method to which an initial position and an end position must be given as an argument. The initial position indicates the position of the first base of the gene, and the stop position indicates the position of the last base. If the stop position is smaller than the initial position, the gene is oriented backwards.
A method __len__ that prints the length of the gene as a result.
A method __repr__ that prints the string representation of a gene of the format "Gene(start, stop)", where start and stop respectively represent the initial and end positions of the gene.
A method __str__ that prints the string representation of a gene of the format "start..stop" for forward genes, and of the format "complement(stop..start)" for backwards genes. Here, start and stop respectively represent the initial and end positions of the gene.
Also, define another class Genome which can be used to represent genome objects. Genome objects must be able to keep the position of their genes, and contain the following methods:
An initializing method to which the length of the genome must be given as an argument.
A method__len__ that prints the length of a genome as a result.
A method addGene to which a gene object must be given as an argument. By calling this method (multiple times), the positions of the genes on the genome can be given. The method must print an AssertionError with the message invalid coordinate if the initial or end position of the given gene object are not within the boundaries of the genome.
A method density that prints the genome density as a floating point value. This method has an optional Boolean parmeter overlap with standard value True. If the value False is given as an argument to the parameter overlap, the genome density must be calculated in the naive way, without taking into account that the genes may overlap. If the value True is given as an argument to the parameter overlap, the genome density must be calculating with the overlaps taken into account.
Example
>>> gene1 = Gene(3309, 4264)
>>> len(gene1)
956
>>> gene1
Gene(3309, 4264)
>>> print(gene1)
3309..4264
>>> gene2 = Gene(14675, 14151)
>>> len(gene2)
525
>>> gene2
Gene(14675, 14151)
>>> print(gene2)
complement(14151..14675)
>>> hiv = Genome(9719)
>>> len(hiv)
9719
>>> hiv.addGene(Gene(1, 634))
>>> hiv.addGene(Gene(790, 2292))
>>> hiv.addGene(Gene(2085, 5096))
>>> hiv.addGene(Gene(5041, 5619))
>>> hiv.addGene(Gene(5559, 5850))
>>> hiv.addGene(Gene(5831, 6045))
>>> hiv.addGene(Gene(5970, 6045))
>>> hiv.addGene(Gene(6062, 6310))
>>> hiv.addGene(Gene(6225, 8795))
>>> hiv.addGene(Gene(8379, 8424))
>>> hiv.addGene(Gene(8379, 8653))
>>> hiv.addGene(Gene(8797, 9417))
>>> hiv.addGene(Gene(9086, 9719))
>>> hiv.density()
98.2302706039716
>>> hiv.density(overlap=False)
110.16565490276777
>>> hiv.addGene(Gene(8888, 9999))
Traceback (most recent call last):
AssertionError: invalid coordinate