Jumping genes
Different regions of the genome evolve in different ways. In coding regions, where any significant change can be lethal to the organism, the most common source of variation is point mutations. In non-coding regions (introns or intergenic spacers), we find a different situation: entire intervals can easily be duplicated, inserted, deleted, or reversed.
During the 1940s and 1950s, Barbara McClintock discovered the effect of genetic transposition in maize plants and demonstrated that many genome rearrangements are caused by regions called transposons, or relatively short intervals of DNA that can change their location within the genome. For the discovery of these "jumping genes", McClintock was awarded the Nobel prize for Physiology or Medicine in 1983.
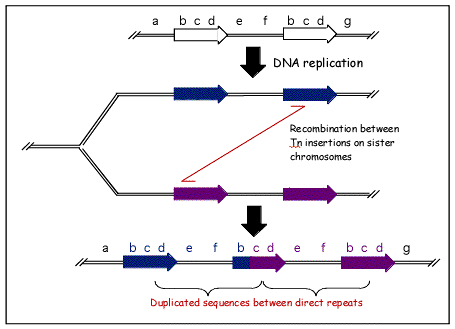
The above figure illustrates the mechanism of transposon duplication during recombination. In this figure, two transposons are turned into three, and any genes located between the two original occurrences of the transposon will be duplicated. This process can also work in the opposite direction to delete genes. Both duplication and deletion can prove harmful to the organism, although deletion is more likely to be harmful.
As with any genomic region, transposons may undergo point mutations and wind up not being completely identical: they only need to be similar enough to facilitate recombination. However, you cannot find multiple repeats in the strings by using the common method for local alignment, as it will find only a single best match.
For example, if you try to locally align the strings AAAATTTTT and TTTTTAAAA, the following optimal local alignment will be returned:
001 1 TTTTT 5
|||||
002 5 TTTTT 9
To find multiple similar regions in two genetic strings simultaneously, we will need to apply a technique called suboptimal alignment, which can obtain more than one different local alignment. When aligning AAAATTTTT and TTTTTAAAA, we should also find the region AAAA of similarity:
001 6 AAAA 9
||||
002 1 AAAA 4
To obtain multiple alternative matches via suboptimal alignment, we can use the program Lalign. Lalign has input and output parameters similar to those of Needle and Water (the "+5/-4" matrix is equivalent to DNAfull), but the user can limit the number of alignments reported by changing the "expectation threshold", which is the maximum number of times a match is expected to occur by random chance. A higher expectation threshold will return more disjoint regions of similarity in the two input strings.
Assignment
The Lalign program for finding multiple alternative matches via suboptimal alignment is available here.
Write a function occurrences that takes two DNA strings
$$s$$ and $$t$$ that share some short inexact repeat $$r$$ of 32-40 bp. By
"inexact" we mean that $$r$$ may appear with slight modifications (each
repeat differs by $$\leq 3$$ changes/indels). The function must return a
tuple containing two integers, indicating the total number of occurrences
of $$r$$ as a substring of $$s$$ and $$t$$ respectively.
Example
In the following interactive sessions, we assume the FASTA files data01.fna, data02.fna and data03.fna to be located in the current directory.
>>> from Bio import SeqIO >>> occurrences(*SeqIO.parse('data01.fna', 'fasta')) (2, 2) >>> occurrences(*SeqIO.parse('data02.fna', 'fasta')) (1, 1) >>> occurrences(*SeqIO.parse('data03.fna', 'fasta')) (3, 6)