A single-nucleotide polymorphism (abbreviation: SNP; pronunciation: snip) is a DNA sequence variation — a polymorphism — occuring when a single nucleotide (A, C, G or T) in the genome (or another shared sequence) differs between two members of a biological species or paired chromosomes in a human. For example, the two sequenced DNA fragments from different individuals, AAGCCTA to AAGCTTA, contain a difference in a single nucleotide at the 5th position. In this case we say that there are two alleles.
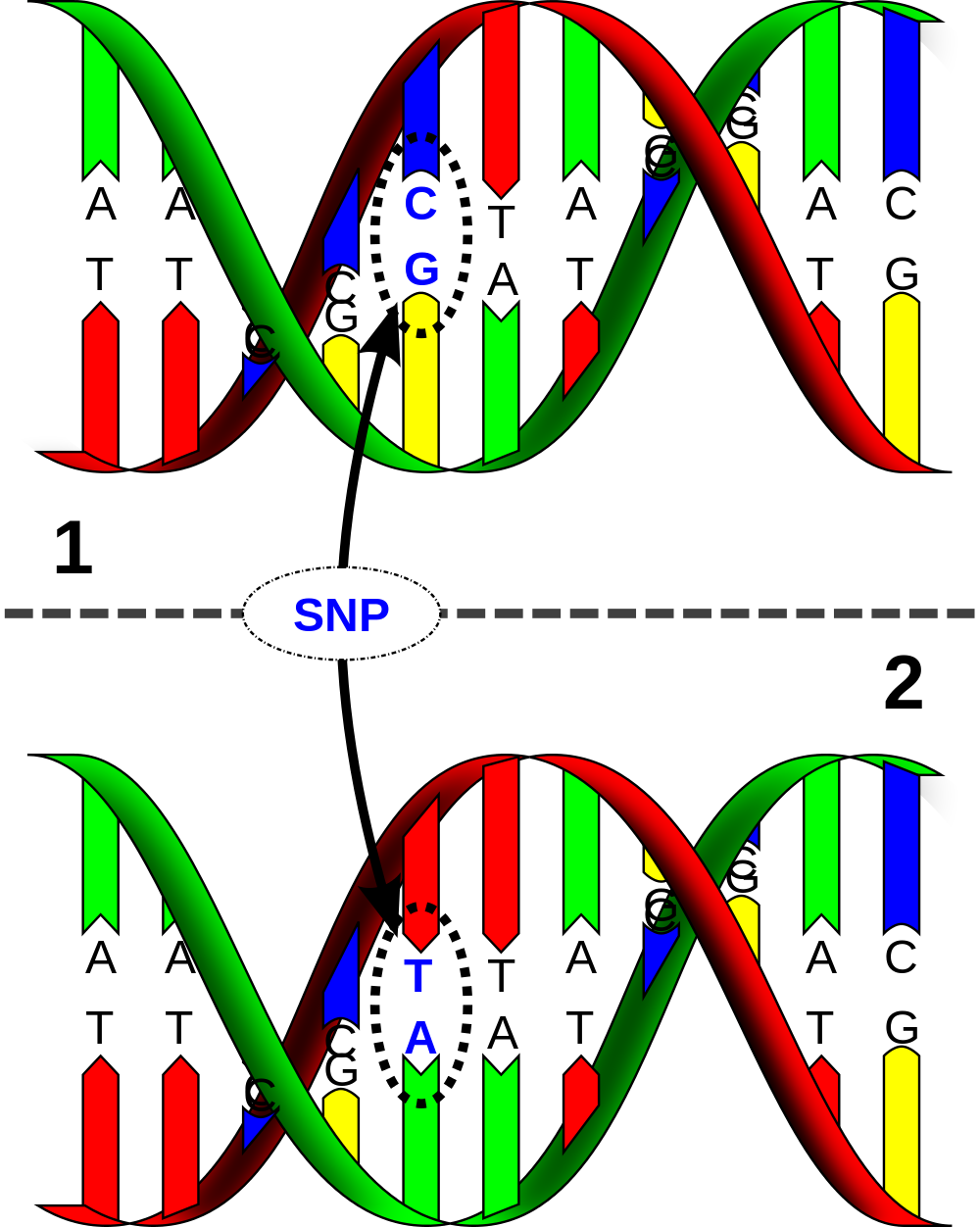
These genetic variations underlie differences in our susceptibility to certain diseases. The severity of illness and the way our body responds to treatments are also manifestations of genetic variations. For example, a single mutation in the APOE (apolipoprotein E1) gene is associated with a higher risk for Alzheimer disease.
Assignment
In this exercise, DNA sequences are represented as strings that only contain the upper case letters A, C, G and T, representing the individual nucleotides.
Write a function SNP that takes two DNA sequences as its arguments. In case these sequences have the same length and only differ at a single location, the function must return a tuple containing three elements. The first element gives the position of the nucleotide that differs between both sequences (positions are indexed from 0). The second and third element are the nucleotides occuring at that position in respectively the first and the second DNA sequence. Otherwise, the function should return the value None.
Searching for SNPs is usually done by scanning the entire genome (or a long DNA fragment) of an individual to compare it with a read (a shorter DNA fragment) of another individual. A SNP is found at a particular position in the genome if all corresponding nucleotides of the read are the same except for a single one. The position of the diverging nucleotide is then used as the position of the SNP. Of course it is possible that multiple SNPs are found when scanning the genome against a single read. An illustration of this is shown in the following picture.
Use the function SNP to write a function SNPs, that takes two arguments: the DNA sequence of a genome and the DNA sequence of a read. The function must return a list containing the positions of all SNPs that are found when comparing the genome against the read. Indexing of the genome positions starts at 0 and the returned list should contain the SNP positions in increasing order.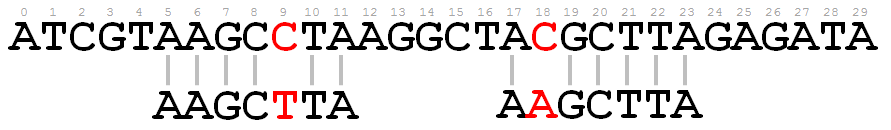
When the read sequence AAGCTTA is mapped onto the genome fragment ATCGTAAGCCTAAGGCTACGCTTAGAGATA, two SNPs are found at positions 9 and 18.
Example
>>> SNP('AAGCCTA', 'AAGCTTA')
(4, 'C', 'T')
>>> SNP('AAGCCTAA', 'AAGCTTA')
>>> SNP('AAGCTTA', 'AAGCTTA')
>>> SNP('AAGCCCA', 'AAGCTTA')
>>> SNPs('AGCTGATAAGCCTAAGCGCT', 'AAGCTTA')
[11]
>>> SNPs('ATCGTAAGCCTAAGGCTACGCTTAGAGATA', 'AAGCTTA')
[9, 18]
>>> SNPs('AAGCCTAAGCCTA', 'AAGCTTA')
[4, 10]