In 2020, Pfizer and BioNTech developed a vaccine against the SARS-CoV-2 virus. It contains genetic material very similar to that of the virus's famous Spike protein: the protein responsible for creating the characteristic club-shaped spikes that project from the surface of coronaviruses.
Through clever chemical processes, the vaccine manages to get this genetic material into specific cells of the human body. These cells then dutifully start producing large quantities of SARS-CoV-2 Spike proteins. When our immune system senses these Spike proteins, it develops a powerful response against the Spike protein and its production process.
Assignment
A peptide is a short stretch of genetic material that is made up of a sequence of building blocks. Each building block is represented by one character (str), so that the peptides can be represented as strings (str). Different characters represent different types of building blocks. No distinction is made between uppercase and lowercase letters.
There are four types of building blocks (called bases) in DNA and RNA. DNA bases are represented by the letters A, C, G and T. In RNA, base T is replaced by base U. This immediately tells us how DNA is transcribed into RNA (and vice versa). In proteins there are 21 types of building blocks (called amino acids), which are represented by 20 letters and an asterisk (*; technically not an amino acid but a stop codon).
A DNA or RNA peptide can describe how proteins are made. To do so, the peptide is split into codons: a sequence of three consecutive bases. As a result, there are $$4^3 = 64$$ different codons. Since each codon corresponds to one amino acid, there must be multiple codons that correspond to the same amino acid. Such codons are called synonyms.
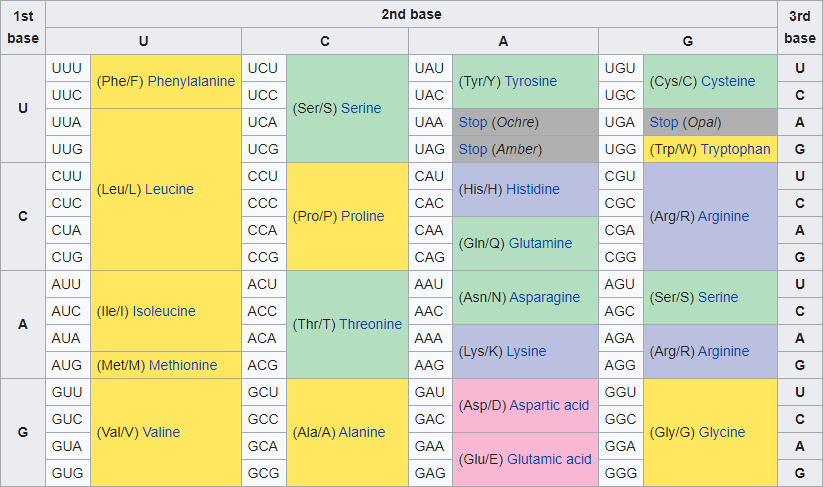
A genetic code describes how codons are converted to amino acids. Different genetic codes (with slight variations) are used by different groups of organisms. A genetic code is stored in a text file in the following format. The first line contains a header (and can therefore be ignored). This is followed by 64 lines, each describing the conversion of a codon to an amino acid using for comma-separated fields: i) codon, ii) amino acid name, iii) three-letter amino acid abbreviation and iv) one-letter amino acid abbreviation. This is, for example, how the standard genetic code is stored:
Codon,Full Name,Abbreviation (3 Letter),Abbreviation (1 Letter) TTT,Phenylalanine,Phe,F TTC,Phenylalanine,Phe,F TTA,Leucine,Leu,L … TAA,Termination (ochre),Ter,* TAG,Termination (amber),Ter,* … GGC,Glycine,Gly,G GGA,Glycine,Gly,G GGG,Glycine,Gly,G
A file describing a genetic code may use either DNA or RNA codons. The codons can be written in uppercase or lowercase.
The Pfizer/BioNTech vaccine uses a technique called codon optimization, where each codon of a peptide is replaced by a synonym containing as many C's and G's as possible. As a result, the peptide still describes the same protein, but it turns out that RNA with a higher amount1 of C's and G's is converted more efficiently into proteins2. This can already be seen, for example, if we compare the start of the SARS-CoV-2 Spike protein with the start of the vaccine:
M F V F L V L L P L V S S Q C V
virus: AUG UUU GUU UUU CUU GUU UUA UUG CCA CUA GUC UCU AGU CAG UGU GUU
| | | | | | | | | | | | |
vaccine: AUG UUC GUG UUC CUG GUG CUG CUG CCU CUG GUG UCC AGC CAG UGU GUU
M F V F L V L L P L V S S Q C V
All mutations are synonymous and all but one introduce an extra C or G in the RNA of the vaccine. This is what gets us to a 95% efficient vaccine. Your task:
-
Write a function genetic_code that takes the location (str) of a text file describing a genetic code $$\mathcal{C}$$. The function must return a dictionary (dict) that maps each DNA codon (str; in uppercase) onto its corresponding amino acid (str; one-letter abbreviation; in uppercase). This is called the dictionary representation of genetic code $$\mathcal{C}$$.
-
Write a function inverse_genetic_code that takes the dictionary representation of a genetic code $$\mathcal{C}$$. The function must return a dictionary (dict) that maps each amino acid (str) onto a set with all synonymous DNA codons (str) corresponding to the amino acid according to genetic code $$\mathcal{C}$$.
-
Write a function synonyms that takes two arguments: i) a codon $$c$$ (str; DNA or RNA; uppercase or lowercase) and ii) the dictionary representation of a genetic code $$\mathcal{C}$$. The function must return a set with all synonyms (str; in uppercase) of codon $$c$$ according to genetic code $$\mathcal{C}$$ (including the codon itself). The function also has an optional parameter RNA that may take a Boolean value (bool) and determines whether the synonyms are returned as RNA codons (True) or as DNA codons (False). If no value is explicitly passed to parameter RNA, it is considered True if at least one U or u appears in codon $$c$$ and False if this is not the case.
-
Write a function codon_optimization that takes a collection (list, tuple or set) of codons (str; DNA or RNA; uppercase or lowercase). The function must return the codon (str) from the collection that contains the most C's and G's. If there are multiple codons in the collection that contain the most C's and G's, the codon that comes alphabetically first must be taken (without making a distinction between uppercase and lowercase letters). This is called the optimal codon among the collection of codons.
-
Write a function peptide_optimization that takes two arguments: i) a peptide $$p$$ (str; DNA or RNA; uppercase or lowercase) and ii) the dictionary representation of a genetic code $$\mathcal{C}$$. The function must return the optimized version (str; in uppercase) of peptide $$p$$, in which each codon $$c$$ is replaced by the optimal codon among the synonyms of codon $$c$$ according to genetic code $$\mathcal{C}$$. If peptide $$p$$ ends with one or two bases that are not part of a codon, they remain unchanged in the optimized version. The function also has an optional parameter RNA that may take a Boolean value (bool) and determines whether the optimized version is returned as an RNA peptide (True) or as a DNA peptide (False). If no value is explicitly passed to parameter RNA, it is considered True if at least one U or u appears in peptide $$p$$ and False if this is not the case.
Note
A genetic code can be described in a text file with DNA or RNA codons, that can in uppercase or lowercase. However, the dictionary representation of a genetic code always uses DNA codons in uppercase, and also has the amino acid letters always in uppercase.
Example
In the following interactive session we assume the text file genetic_code.txt3 to be located in the current directory.
>>> code = genetic_code('genetic_code.txt4')
>>> code['TCT']
'S'
>>> code['TCA']
'S'
>>> code['ATG']
'M'
>>> inverse_code = inverse_genetic_code(code)
>>> inverse_code['S']
{'TCA', 'TCC', 'TCT', 'AGC', 'TCG', 'AGT'}
>>> inverse_code['M']
{'ATG'}
>>> inverse_code['*']
{'TGA', 'TAA', 'TAG'}
>>> synonyms('TCT', code)
{'TCC', 'AGT', 'TCT', 'AGC', 'TCA', 'TCG'}
>>> synonyms('atg', code, RNA=True)
{'AUG'}
>>> synonyms('UGA', code)
{'UAA', 'UAG', 'UGA'}
>>> codon_optimization(['tca', 'tcc', 'tct', 'agc', 'tcg', 'agt'])
'agc'
>>> codon_optimization({'ATG'})
'ATG'
>>> codon_optimization(('UAA', 'UAG', 'UGA'))
'UAG'
>>> peptide_optimization('ATGTTTGTTTTTCTTGTTTTATTGCCACTAGTCTCTAGTCAGTGTGTT', code)
'ATGTTCGTCTTCCTCGTCCTCCTCCCCCTCGTCAGCAGCCAGTGCGTC'
>>> peptide_optimization('AUGUUUGUUUUUCUUGUUUUAUUGCCACUAGUCUCUAGUCAGUGUGUU', code, RNA=True)
'AUGUUCGUCUUCCUCGUCCUCCUCCCCCUCGUCAGCAGCCAGUGCGUC'