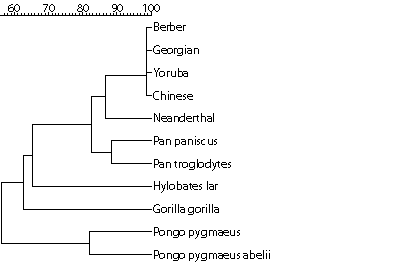
The discovery of skeletons of the Neanderthal (Homo sapiens neanderthalensis) has raised many questions about the human origin in different parts of Europe, including the search for our relationship to this species. Such questions about the origin of humans and primates are normally answered by studying the mitochondrial DNA, and in particular the hypervariable regions within them. These regions show high variability within the human race, making them ideal for the study of relationships between individuals. The mitochondrial DNA exhibits two hypervariable regions, which are designated as HVR-I and HVR-II, respectively.
In order to determine the similarity (measure of kindship) of DNA sequences, first of all an alignment of all investigated sequences is constructed. In such a sequence alignment the corresponding parts of two or more sequences are placed underneath each other. Below is an example of such a sequence alignment between two sequences.
CTG-GGG--GGTGTAC || ||| | || | CTACGGG---GCGTCC
Here, the corresponding base pairs (matches) are indicated by vertical lines between the base pairs, and holes are represented by hyphens (-) within the sequences (causing the aligned sequences to always have the same length). Errors (mismatches) can be explained by point mutations and holes (gaps) by insertions or deletions.
On the basis of a sequence alignment, the similarity between two sequences can be calculated in the following way. For corresponding base pairs, a score of +1 is awarded, for errors a score of 0 and for holes a score of -1. Positions where both sequences have a gap will not be taken into account. The scores for each position of the alignment are then added together and divided by the number of positions brought into account. In the above example, there are 9 pairs of corresponding base pairs, 3 errors, 2 holes, and 14 of the 16 positions are taken into account (two positions exhibit a hole in both sequences). The similarity of these two sequences equals \[ \frac{9 \times (+1) + 3 \times 0 + 2 \times (-1)}{14} = 0.5 \]
Assignment
-
Write a function score, that returns the corresponding score for two given base pairs (that must be passed to the function as argument) based on the values from the table below:
type example score match A en A +1 mismatch A en G 0 1 hole A en - -1 2 holes - en - 0 -
Use the function score to write a function similarity that calculates the corresponding similarity for two given DNA sequences of a sequence alignment as was described above. This function should return the value of the similarity. For example, the function should return the value 0.5 for the DNA sequences "CTG-GGG--GGTGTAC" and "CTACGGG---GCGTCC" from the example above.
Note: If you have implemented the similarity function properly it will be used to establish a phylogenetic tree on the basis of the mitochondrial DNA of some primates. Based on this result, can you find out whether modern humans evolved from Neanderthals?
Example
>>> similarity('CTG-GGG--GGTGTAC', 'CTACGGG---GCGTCC')
0.5
>>> similarity('CTG-GGG---GTGTAC', 'CTACGGG---GCGTCC')
0.6153846153846154
>>> similarity('CTG-GGG--GGTGTAC', 'CTACGGG---GCGTCA')
0.42857142857142855Epilogue: Altamura Man
About 150,000 years ago, a Neanderthal man was exploring the Lamalunga Cave in southern Italy when he fell into a sinkhole. Too badly injured to climb out again, he died of dehydration or starvation. Over the ensuing centuries, water running down the cave walls gradually incorporated the man's bones into concretions of calcium carbonate. Undisturbed by predators or weather, they lay in an immaculate state of preservation until cave researchers finally discovered them in 1993.
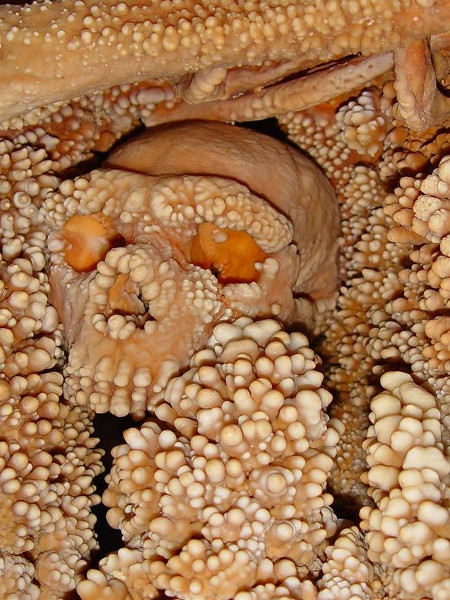
This is a great boon for paleoanthropologists — Altamura Man is one of the most complete Paleolithic skeletons ever discovered in Europe — but there's a downside: the bones have become so deeply involved in their matrix of limestone that no one has found a way to remove them without destroying them. So, for now, all research must be carried out in the cave.