The Russian astrophysicist George Gamow is seen as the father of the Big Bang theory. A title he owes to his prediction of the cosmic microwave background radiation. Gamow was a creative thinker who felt quite at home taking a sidestep into another discipline. His contribution to cracking the genetic code is seen as "perhaps the last example of amateurism in scientific work on grand scale". After all, it was Gamow's idea that the base sequence in DNA might be the code for protein synthesis.
In the spring of 1953, Francis Crick and James Watson decoded the structure of deoxyribonucleic acid (DNA). They discovered that DNA — the molecular basis of heredity — consists of two intertwined strands that run in opposite directions. Each strand is a long molecule comprising a string of sugars, phosphate groups, and one of the following four bases: adenine (A), thymine (T), cytosine (C) or guanine (G). The burning question was soon raised: "How is the information in DNA converted into the production of amino acids, the building blocks of proteins?".
The first step in finding a solution came from an unexpected quarter. After reading the work of Watson and Crick, George Gamow wrote them a letter in the summer of 1953. He suggested that the base sequence in DNA might be the code for protein synthesis. As a physicist, Gamow's idea took the world of biology by storm. He had changed what had, until then, been seen as a chemical problem into purely a question of information storage and transfer. The underlying chemistry was of secondary importance.
Gamow had reduced the problem to the question: "How can a language of four letters provide a code for 20 amino acids?". It soon became clear that the four different bases had to be grouped in threes — in this context these triplets are often called codons — to make a unique code for each of the 20 amino acids possible. Groups of two only allow for 16 ($$4 \times 4$$) possibilities, while triplets provide 64 ($$4\times 4 \times 4$$) possibilities, which is more than enough.
Gamow himself made the first proposal, what is known as the diamond code. He thought that the protein synthesis occurred directly between the two strands of DNA. The four bases form a space in which an amino acid fits perfectly. Which acid that is, depends on the bases of the four corner points, hence the name diamond. The bases at the left and right corners of the diamond lie on the same strand, separated by a single base. This base and its complement on the opposite strand constitute the top and bottom corners of the diamond (A is complementary to T and C is complementary to G). In essence, Gamow's code was a three-letter code, as the top and bottom corners were complementary, so that only one of the two actually contained information.
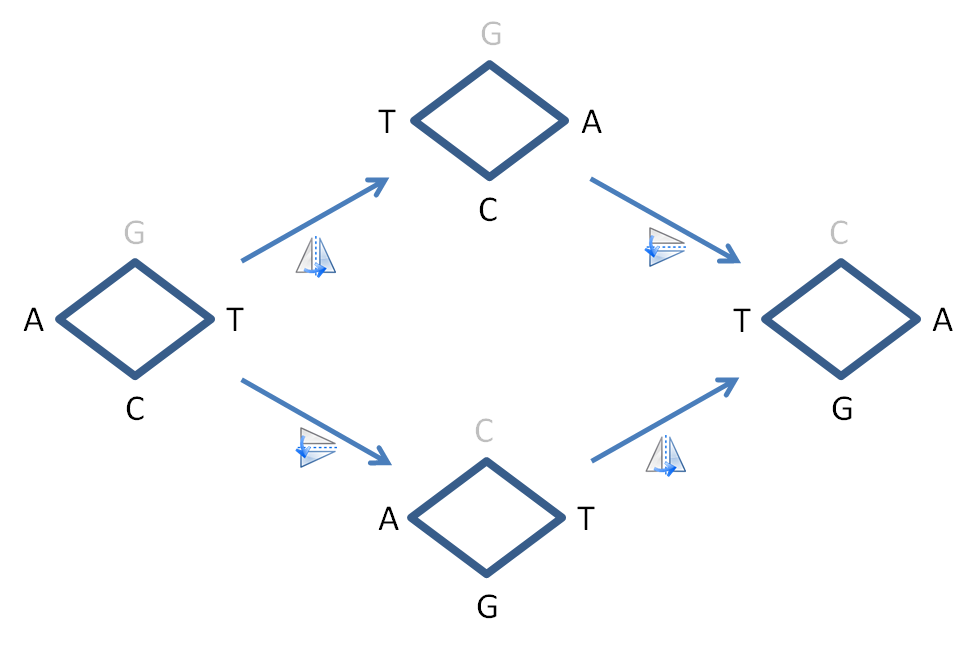
Gamow's diamond was also an overlapping code. Each base was part of three sequential codons. For example, the base sequence ATCGAT consisted of the four codons ATC, TCG, CGA and GAT. Gamow came up with an original solution for the 64 possible codons for only 20 amino acids. He suggested that the diamonds could, as it were, be rotated on both axes without that having any significance. If the ACT codon were rotated on the vertical axis, it would become TCA. Rotating it on the horizontal axis would replace the middle base with its complement, making it AGT. If all these symmetries are fully worked out, you end up with 20 unique combinations. The exact number Gamow was looking for.
Assignment
In this exercise we will represent both DNA and protein sequences as strings that only contain uppercase letters. DNA sequences are limited to the letters A, C, G and T, that represent the possible nucleotides. A codon is a DNA sequence that has three letters. Protein sequences may contain each letter of the alphabet (in practice only 20 letters are used), which now represent the possible amino acids. Your task is to convert DNA sequences into their corresponding protein sequence according to the principles of Gamow's diamond code. Follow these steps to accomplish this task:
-
Write a function canonical that returns the canonical representation of the codon that is passed as an argument to the function. The canonical representation of a given codon is determined by rotating it on the horizontal and/or vertical axis of the diamond representation of the codon. The canonical representation is the alphabetically first ranked of the (up to) four codons that result from these rotations.
-
Use the function canonical to write a function codon2aa that takes a codon as its argument. The function must return a single letter that represents the amino acid corresponding to the given codon. The letter should be determined in the following way:
-
Determine the canonical representation $$b_1b_2b_3$$ of the given codon.
-
Compute \[ p = (w_1 + 4w_2 + 16w_3)\!\!\!\!\mod 25 \] In this, $$w_i$$ corresponds to the value of the nucleotide $$b_i$$ ($$1 \leq i \leq 3)$$, with nucleotide G having value 0, T having value 1, C having value 2 and A having value 3.
-
The value $$p$$ gives the position in the alphabet of the letter of the amino acid that corresponds to the given codon. Positions in the alphabet are indexed from zero, so that A is at position 0, B at position 1, C at position 2, …
-
-
Use the function codon2aa to write a function dna2protein. This function should be passed a DNA sequence that has at least three nucleic acids. The function must return the corresponding protein sequence according to Gamow's diamond code. We remind you once more that this is an overlapping code.
Example
>>> canonical('ACT')
'ACT'
>>> canonical('CGC')
'CCC'
>>> canonical('GTC')
'CAG'
>>> len(set([canonical(a + b + c) for a in 'ACGT' for b in 'ACGT' for c in 'ACGT']))
20
>>> codon2aa('ACT')
'C'
>>> codon2aa('CGC')
'R'
>>> codon2aa('GTC')
'O'
>>> len(set([codon2aa(a + b + c) for a in 'ACGT' for b in 'ACGT' for c in 'ACGT']))
20
>>> dna2protein('ATCGAT')
'WYSD'
>>> dna2protein('CCCTCCATCTAGTGCGTGTTCTGTCCGAAGGTATGTCATATCAC')
'RBVBSFWAWDCMBIBMADFAOAOBKSPPLYPEPAOCFNEWCV'
>>> dna2protein('ATTTAACGAATCTACCCGGAGTGGCAACTCAGGAGGACTCTTG')
'GEGGWLSPGWAWFSRKKLMCMYKLWWCVCOLLMLLOCVAFD'Epilogue
By the time of his trip to biology Gamow had already turned fifty and had a long academic career behind him. He was most famous for his work on quantum mechanics and nuclear physics. Gamow came to some remarkable predictions for his time, simply by applying accepted laws of nature to unusual situations. As such, he predicted in 1948 that there should be a measurable amount of cosmic background radiation if the universe had a hot and compact beginning. Almost twenty years later, the existence of cosmic background radiation was indeed confirmed experimentally.
After he had proposed the diamond code, however, Gamow soon realised that this code was not the correct solution. This was just as well, since it was very sensitive to mutations. With an overlapping code, mutation of one base can impact three successive amino acids. In the meantime, others had become convinced that protein synthesis did not directly occur in DNA, but that ribonucleic acid (RNA) acted as an intermediary. RNA is very similar to DNA, but consists of a single strand of sugars, phosphates, and bases. It also contains the base uracil (U) instead of thymine.
Although his diamond code proved incorrect, Gamow was not ready to throw in the towel. He had formed an informal group of scientists who were more or less involved in addressing the code problem. His RNA Tie Club had 20 regular members, one for each amino acid, and four honorary members, one for each base. Gamow himself was alanine (ALA), Watson was proline (PRO), and Crick tyrosine (TYR). The other members were mainly biologists, like Max Delbrück (tryptophan) and Erwing Chargaff (lysine), but Gamow did not repudiate his own background, enlisting a number of leading physicists, including Edward Teller (leucine) and Richard Feynman (glycine). Each member received a specially designed tie bearing a double helix and a tiepin with the acronym of their own personal amino acid. The RNA Tie Club's official notepaper carried the motto "Do or die, or don't try".
After the diamond code, Gamow came up with two alternative codes, one of which he devised together with Feynman. Even Teller, a nuclear physicist pur sang, took the time to propose an interesting scheme, in which each amino acid was encoded by two bases and the preceding amino acid. In 1957, Syndey Brenner (valine) abruptly put a stop to all overlapping codes, when they proved incompatible with his analysis of the sequence of amino acids in a number of proteins.
That same year, Crick launched an ingenious non-overlapping code. He claimed that there was only one way in which the base sequence could be read. Imagine that the base sequence AGACGAUUA coded for AGA, CGA and UUA. According to Crick, the triplets of the other two overlapping codes were "nonsense codons", with no significance at all. In this case, therefore, GAC and GAU on the one hand, and ACG and AUU on the other hand, would be nonsense codons. Crick's code was incorrect, but was called "the most elegant biological theory ever to be proposed and proved wrong".
With hindsight, the RNA Tie Club had been too focused on finding a neat explanation of why there are 64 codes for only 20 amino acids. They were brought down to earth in 1961 when Marshall Nirenberg and Heinrich Matthaei — neither members of the RNA Tie Club — announced that they were able to produce proteins with artificial RNA. The first RNA they tested was poly-U, a sequence or uracil bases. They discovered that UUU coded for the amino acid phenylalanine. Four years later, the whole coding problem was solved. Compared to the solutions proposed earlier, nature's solution seemed like a rather messy workaround. Some amino acids have only one codon, while others have four, and some even six. Although the real solution was less refined mathematically than his own idea, Gamow admitted that it had one great advantage: it was true.
Resources
-
Sanger F, Tuppy H (1951). The amino acid sequence in the phenylalanyl chain of insulin. I. The identification of lower peptides from partial hydrolysates. Biochemical Journal 49, 463-481. 1
-
Watson JD, Crick FHC (1953). A structure of deoxyribose nucleic acid. Nature 171, 737-738. 2
-
Gamow G (1954). Possible relation between deoxyribonucleic acid and protein structures. Nature 173, 318. 3
-
Brenner S (1957). On the impossibility of all overlapping triplet codes in information transfer from nucleic acid to proteins. Proceedings of the National Academy of Sciences of the USA 43, 687-694. 4
-
Crick FHC, Griffith JS, Orgel LE (1957). Codes without commans. Proceedings of the National Academy of Sciences of the USA 43, 416-421. 5
-
Marshall NW, Matthaei J (1961). The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proceedings of the National Academy of Sciences of the USA 47, 1588-1602. 6
-
Hayes B (1998). The invention of the genetic code. American Scientist 86, 814. 7
-
Patel A (2001). Why genetic information processing could have a quantum basis. Journal of Biosciences 26(2), 145-151. 8
-
Sarabhai A (2003). After DNA at the MRC. Journal of Biosciences 28(6), 665-669. 9
-
Freeland SJ, Hurst LD (2004). Evolution encoded. Scientific American 290(4), 84-91. 10