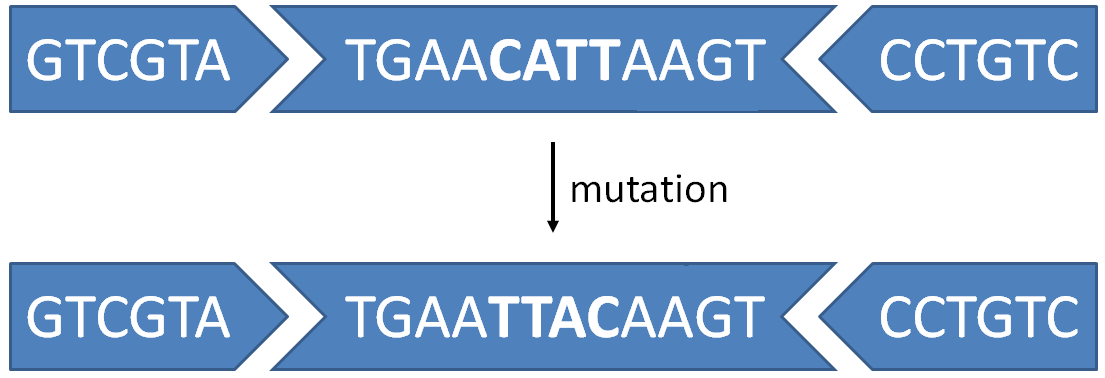
DNA sequences can be represented as strings that only contain the letters A, C, G and T. One particular type of mutation that is observed in DNA, occurs when a particular subsequence of the DNA is completely inverted during the replication process. Usually such reversals occur in between what is called inverted pairs. For example, if the pattern TGAA is followed further downstream by the pattern AAGT, it is possible that the DNA segment that is enclosed in between these two patterns will be inverted and reattached, because binding regions at both ends are locally the same.
Assignment
Write a function mutation that is passed a DNA sequence and a sequence pattern (both must be passed as string arguments to the function) and returns the mutated DNA sequence. The function should do this by finding the leftmost position of the pattern within the sequence, and next occurrence of the inverted pattern. The mutated DNA sequence is the result by reverting the segment in between the inverted pair. The function must return the original (non-mutated) DNA sequence in case the pattern does not occur in the original DNA sequence, or if it is not followed further downstream by the inverted pattern.
Example
>>> mutation('GTCGTATGAACATTAAGTCCTGTC', 'TGAA')
'GTCGTATGAATTACAAGTCCTGTC'
>>> mutation('GTCGTATGAACATTAAGTCCTGTC', 'GTC')
'GTCCTGAATTACAAGTATGCTGTC'
>>> mutation('GTCGTATGAACATTAAGTCCTGTC', 'TCG')
'GTCGTATGAACATTAAGTCCTGTC'The following table illustrates how the results in the above example were found, with inverted pairs being underlined.
| SEQUENCE | PATTERN | RESULT |
|---|---|---|
| GTCGTATGAACATTAAGTCCTGTC | TGAA | GTCGTATGAATTACAAGTCCTGTC |
| GTCGTATGAACATTAAGTCCTGTC | GTC | GTCCTGAATTACAAGTATGCTGTC |
| GTCGTATGAACATTAAGTCCTGTC | TCG | GTCGTATGAACATTAAGTCCTGTC |