The heights we have been looking at are not the original heights reported by students. The original reported heights are also included in the dslabs package and can be loaded like this:
library(dslabs)
data("reported_heights")
Height is a character vector so we create a new column with the numeric version:
reported_heights <- reported_heights %>%
mutate(original_heights = height, height = as.numeric(height))
#> Warning: Problem with `mutate()` input `height`.
#> x NAs introduced by coercion
#> ℹ Input `height` is `as.numeric(height)`.
#> Warning in mask$eval_all_mutate(dots[[i]]): NAs introduced by coercion
Note that we get a warning about NAs. This is because some of the self reported heights were not numbers. We can see why we get these:
reported_heights %>% filter(is.na(height)) %>% head()
#> time_stamp sex height original_heights
#> 1 2014-09-02 15:16:28 Male NA 5' 4"
#> 2 2014-09-02 15:16:37 Female NA 165cm
#> 3 2014-09-02 15:16:52 Male NA 5'7
#> 4 2014-09-02 15:16:56 Male NA >9000
#> 5 2014-09-02 15:16:56 Male NA 5'7"
#> 6 2014-09-02 15:17:09 Female NA 5'3"
Some students self-reported their heights using feet and inches rather than just inches. Others used centimeters and others were just trolling. For now we will remove these entries:
reported_heights <- filter(reported_heights, !is.na(height))
If we compute the average and standard deviation, we notice that we obtain strange results. The average and standard deviation are different from the median and MAD:
reported_heights %>%
group_by(sex) %>%
summarize(average = mean(height), sd = sd(height),
median = median(height), MAD = mad(height))
#> `summarise()` ungrouping output (override with `.groups` argument)
#> # A tibble: 2 x 5
#> sex average sd median MAD
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Female 63.4 27.9 64.2 4.05
#> 2 Male 103. 530. 70 4.45
This suggests that we have outliers, which is confirmed by creating a boxplot:
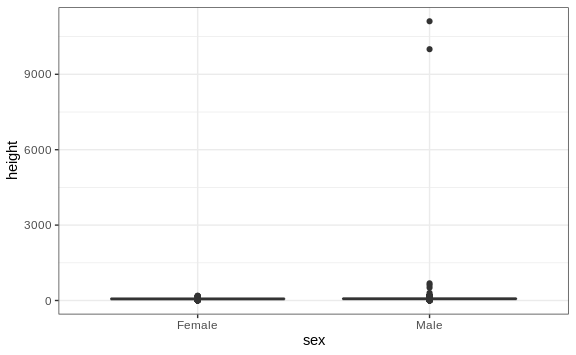
We can see some rather extreme values. To see what these values are, we
can quickly look at the largest values using the arrange function:
reported_heights %>% arrange(desc(height)) %>% top_n(10, height)
#> time_stamp sex height original_heights
#> 1 2014-09-03 23:55:37 Male 11111 11111
#> 2 2016-04-10 22:45:49 Male 10000 10000
#> 3 2015-08-10 03:10:01 Male 684 684
#> 4 2015-02-27 18:05:06 Male 612 612
#> 5 2014-09-02 15:16:41 Male 511 511
#> 6 2014-09-07 20:53:43 Male 300 300
#> 7 2014-11-28 12:18:40 Male 214 214
#> 8 2017-04-03 16:16:57 Male 210 210
#> 9 2015-11-24 10:39:45 Male 192 192
#> 10 2014-12-26 10:00:12 Male 190 190
#> 11 2016-11-06 10:21:02 Female 190 190
The first seven entries look like strange errors. However, the next few look like they were entered as centimeters instead of inches. Since 184 cm is equivalent to six feet tall, we suspect that 184 was actually meant to be 72 inches.
We can review all the nonsensical answers by looking at the data considered to be far out by Tukey:
whisker <- 3*IQR(reported_heights$height)
max_height <- quantile(reported_heights$height, .75) + whisker
min_height <- quantile(reported_heights$height, .25) - whisker
reported_heights %>%
filter(!between(height, min_height, max_height)) %>%
select(original_heights) %>%
head(n=10) %>% pull(original_heights)
#> [1] "6" "5.3" "511" "6" "2" "5.25" "5.5" "11111"
#> [9] "6" "6.5"
Examining these heights carefully, we see two common mistakes: entries
in centimeters, which turn out to be too large, and entries of the form
x.y with x and y representing feet and inches, respectively, which
turn out to be too small. Some of the even smaller values, such as 1.6,
could be entries in meters.
In the Data Wrangling part of this book we will learn techniques for correcting these values and converting them into inches. Here we were able to detect this problem using careful data exploration to uncover issues with the data: the first step in the great majority of data science projects.