The first step when importing data from a spreadsheet is to locate the file containing the data. Although we do not recommend it, you can use an approach similar to what you do to open files in Microsoft Excel by clicking on the RStudio “File” menu, clicking “Import Dataset”, then clicking through folders until you find the file. We want to be able to write code rather than use the point-and-click approach. The keys and concepts we need to learn to do this are described in detail in the Productivity Tools part of this book. Here we provide an overview of the very basics.
The main challenge in this first step is that we need to let the R functions doing the importing know where to look for the file containing the data. The simplest way to do this is to have a copy of the file in the working directory, in which the importing functions look by default. Once we do this, all we have to supply to the importing function is the filename.
The filesystem
You can think of your computer’s filesystem as a series of nested folders, each containing other folders and files. Data scientists refer to folders as directories. We refer to the folder that contains all other folders as the root directory. We refer to the directory in which we are currently located as the working directory. The working directory therefore changes as you move through folders: think of it as your current location.
The working directory
The working directory is just a file path on your computer that sets the default location of any files you read into R, or save out of R. In other words, a working directory is like a little flag somewhere on a folder in your filesystem.
You can get the full path of your working directory by using the getwd function.
wd <- getwd()
If you need to change your working directory, you can use the function
setwd or you can change it through RStudio by clicking on “Session”.
setwd("path/to/a/folder")
Once the file is copied into the working directory, we can import the data with a simple line of code. Here we use the read_csv function from the readr package, which is part of the tidyverse.
library(tidyverse)
dat <- read_csv("filename.csv")
The data is imported and stored in dat.
Relative and full paths
We could now copy all of our files to the working directory, but sometimes this is just not what we want. Therefor it is possible to import a file by its path rather than only its filename.
The path of a file is a list of directory names that can be thought of as instructions for the computer on what folders to click on, and in what order, to find the file. If these instructions are for finding the file starting from the root directory we refer to it as the full path. If the instructions are for finding the file starting in the working directory we refer to it as a relative path.
Full paths
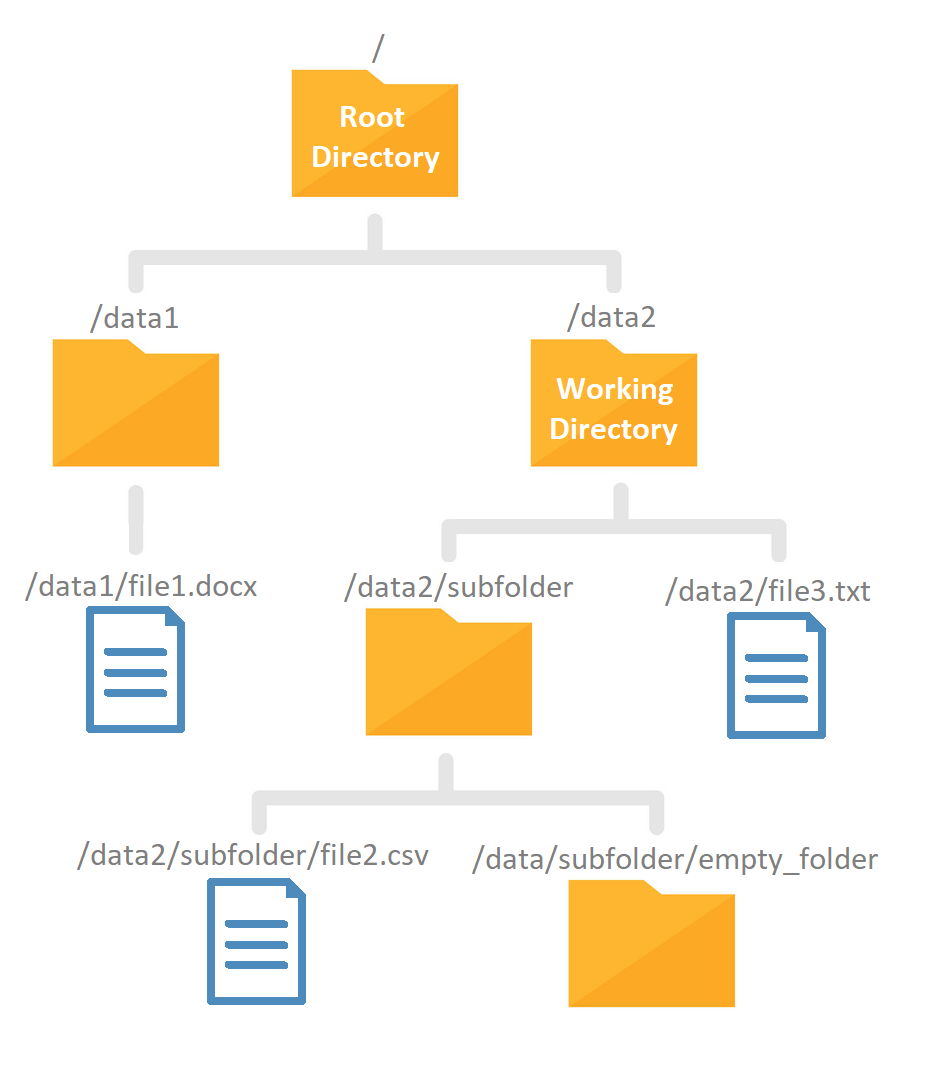
In the illustration above represents a file structure where every folder and file is labeled with it’s absolute path.
The strings separated by slashes are the directory names. The first slash represents the root directory and we know this is a full path because it starts with a slash.
Relative paths
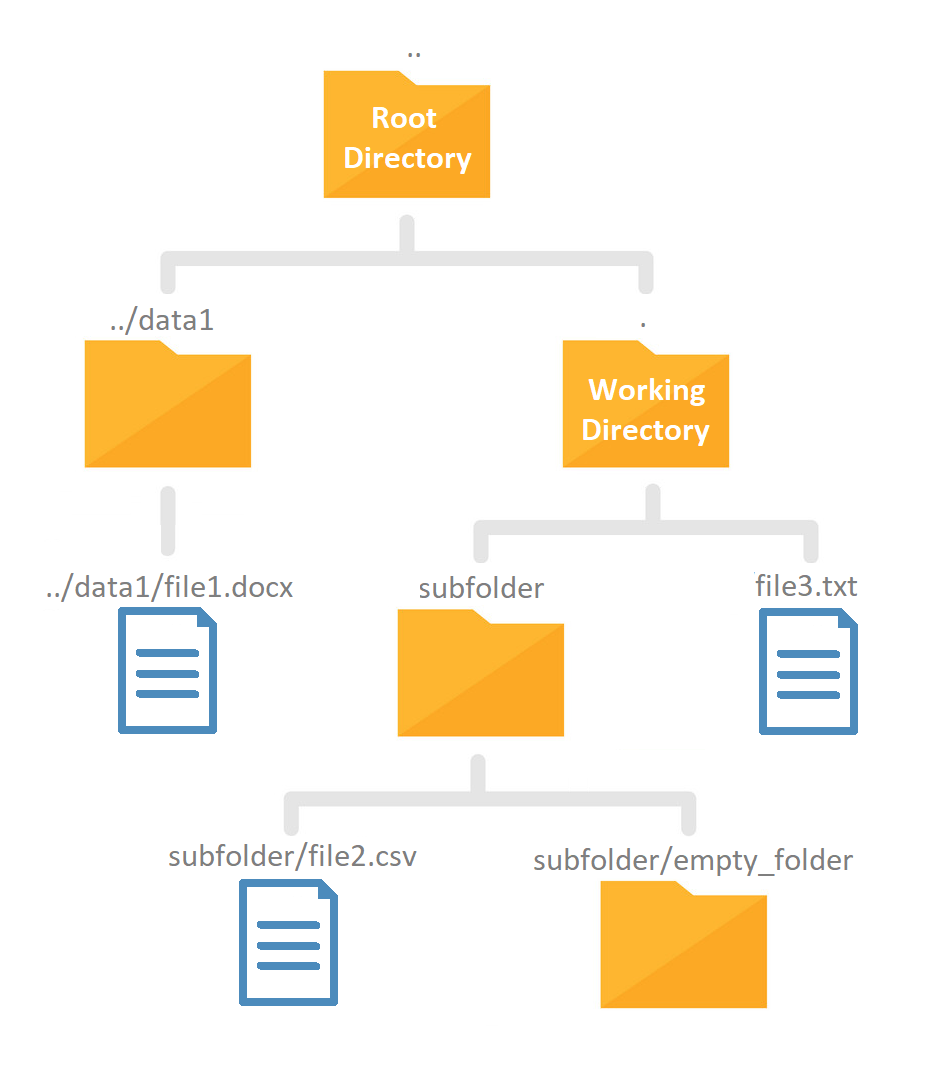
In the illustration above represents a file structure where every folder and file is labeled with it’s relative path.
If the first directory name appears without a slash in front, then the path is assumed to be relative.
These relative paths give us the location of the files or directories if we start in the working directory.
Because every path starts from the working directory you might think that you can only reference folders and files located inside this folder. This is not true, a relative path can acces all folders and files an absolute path can acces. this is possible because of the double dot .. that references the parent directory. For example: if your working directory is empty_folder (from the illustration above) and you want to acces data1 you will need to go up three parent directories and then specify data1: ../../../data1.
Use Relative Paths!!!
We highly recommend only writing relative paths in your code. The reason is that full paths are unique to your computer and you want your code to be portable to any computer system.
Generating path names
In the following example the full path to the murders dataset (from the dslabs package) is constructed by code:
filename <- "murders.csv"
dir <- system.file("extdata", package = "dslabs")
fullpath <- file.path(dir, filename)
The function system.file provides the full path of the folder containing all the files and directories relevant to the package
specified by the package argument. By exploring the directories in
dir we find that the extdata contains the file we want:
dir <- system.file(package = "dslabs")
filename %in% list.files(file.path(dir, "extdata"))
#> [1] TRUE
The system.file function permits us to provide a subdirectory as a
first argument, so we can obtain the fullpath of the extdata directory
like this:
dir <- system.file("extdata", package = "dslabs")
The function file.path is used to combine directory names to produce
the full path of the file we want to import.
fullpath <- file.path(dir, filename)
Copying files using paths
The final line of code we used to copy the file into our home directory
used
the function file.copy. This function takes two arguments: the file to
copy and the name to give it in the new directory.
file.copy(fullpath, "murders.csv")
#> [1] TRUE
If a file is copied successfully, the file.copy function returns
TRUE. Note that we are giving the file the same name, murders.csv,
but we could have named it anything. Also note that by not starting the
string with a slash, R assumes this is a relative path and copies the
file to the working directory.
You should be able to see the file in your working directory and can check by using:
list.files()