Next we add details to modnn that control the fitting algorithm. Here we
have simply followed the examples given in the Keras book. We minimize
squared-error loss as in
The algorithm tracks the mean absolute error on the training data, and on validation data if it is supplied.
modnn %>% compile(loss = "mse",
optimizer = optimizer_rmsprop(),
metrics = list("mean_absolute_error"))
In the previous line, the pipe operator passes modnn as the first argument
to compile(). The compile() function does not actually change the R object
modnn, but it does communicate these specifications to the corresponding
python instance of this model that has been created along the way.
Now we fit the model. We supply the training data and two fitting parameters,
epochs and batch_size. Using 32 for the latter means that at each
step of SGD, the algorithm randomly selects 32 training observations for
the computation of the gradient. Recall from Sections 10.4 and 10.7 that
an epoch amounts to the number of SGD steps required to process \(n\) observations.
Since the training set has \(n = 176\), an epoch is \(176/32 = 5.5\) SGD
steps. The fit() function has an argument validation_data; these data are
not used in the fitting, but can be used to track the progress of the model
(in this case reporting the mean absolute error). Here we actually supply
the test data so we can see the mean absolute error of both the training
data and test data as the epochs proceed. To see more options for fitting,
use ?fit.keras.engine.training.Model.
history <- modnn %>% fit(
x[-testid,], y[-testid], epochs = 1500, batch_size = 32,
validation_data = list(x[testid,], y[testid]))
We can plot the history to display the mean absolute error for the training
and test data. For the best aesthetics, install the ggplot2 package before
calling the plot() function. If you have not installed ggplot2, then the code
below will still run, but the plot will be less attractive.
plot(history)
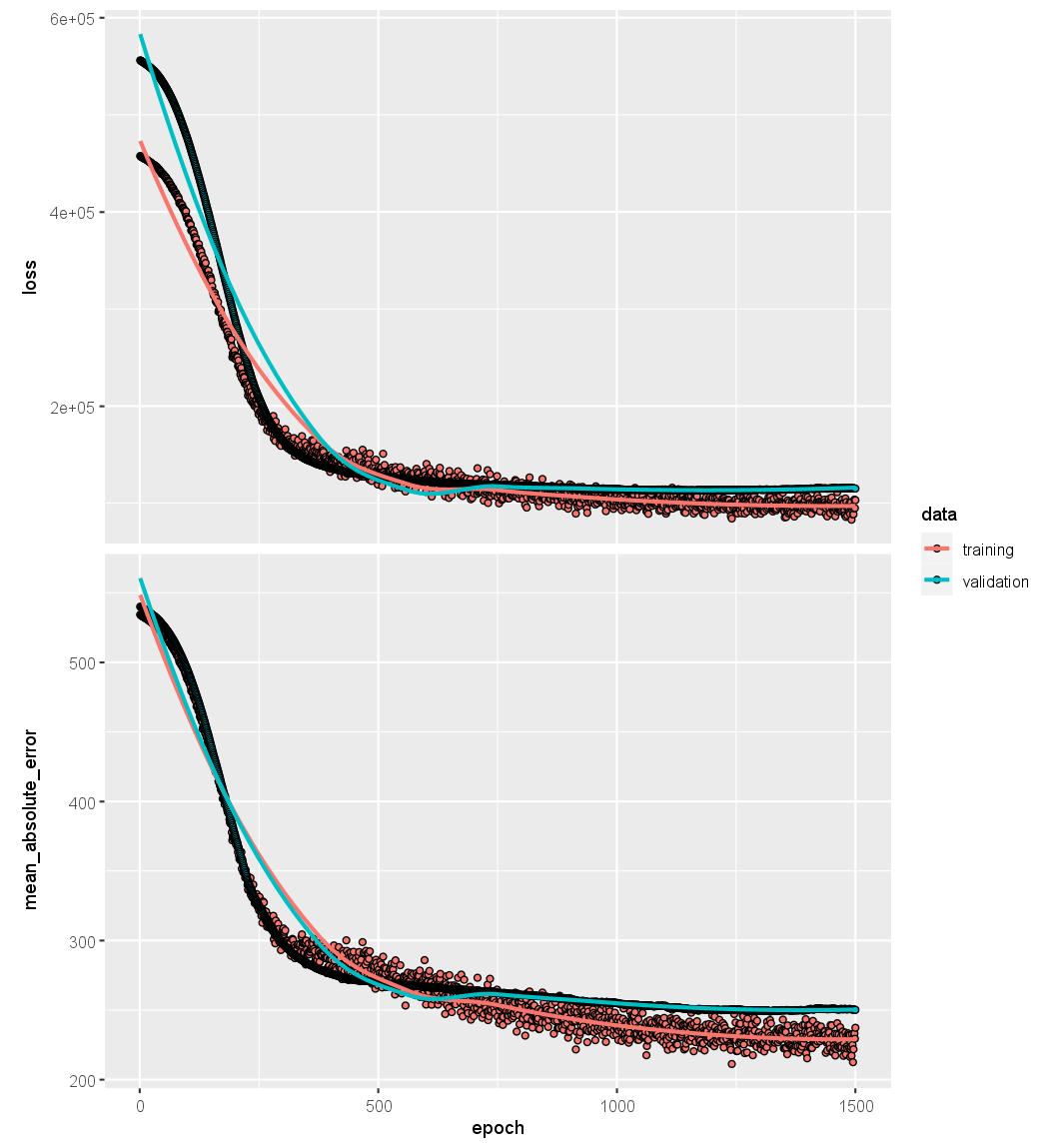
It is worth noting that if you run the fit() command a second time in the
same R session, then the fitting process will pick up where it left off. Try
re-running the fit() command, and then the plot() command, to see!
Finally, we predict from the final model, and evaluate its performance
on the test data. Due to the use of SGD, the results vary slightly with each
fit. Unfortunately the set.seed() function does not ensure identical results
(since the fitting is done in python), so your results will differ slightly.
npred <- predict(modnn, x[testid,])
mean(abs(y[testid] - npred))
[1] 250.1202
Questions
- Compile the
modnnmodel from the previous exercise as follows:- Use the MSE as the loss function
- Use the
optimizer_adam()as you optimizer - Also track the
mean_squared_erroras a metric.
- Fit the neural network on the training data for 1000 epochs and a batch size of 32.
Supply the test data also as
validation_dataso you can monitor the train & test MSE. Store the result inhistory. - Visualize the loss and mean absolute error for the training and validation data in function of the number of epochs.
- Store the predictions on the test data of the neural network in
npred. - Store the MSE of the neural network on the test data in
nmse.