Genes are discontinuous
In "Transcribing DNA into RNA" we mentioned that a strand of DNA is copied into a strand of RNA during transcription, but we neglected to mention how transcription is achieved.
In the nucleus, an enzyme (i.e., a molecule that accelerates a chemical reaction) called RNA polymerase (RNAP) initiates transcription by breaking the bonds joining complementary bases of DNA. It then creates a molecule called precursor mRNA, or pre-mRNA, by using one of the two strands of DNA as a template strand: moving down the template strand, when RNAP encounters the next nucleotide, it adds the complementary base to the growing RNA strand, with the provision that uracil must be used in place of thymine.
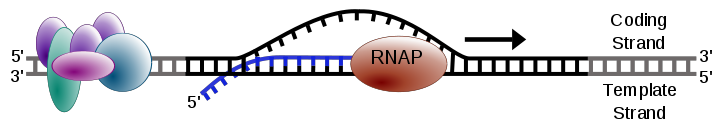
Because RNA is constructed based on complementarity, the second strand of DNA, called the coding strand, is identical to the new strand of RNA except for the replacement of thymine with uracil. See figure below and recall "Transcribing DNA into RNA".
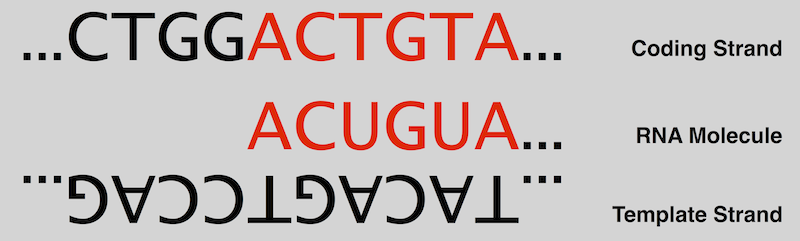
After RNAP has created several nucleotides of RNA, the first separated complementary DNA bases then bond back together. The overall effect is very similar to a pair of zippers traversing the DNA double helix, unzipping the two strands and then quickly zipping them back together while the strand of pre-mRNA is produced.
For that matter, it is not the case that an entire substring of DNA is transcribed into RNA and then translated into a peptide one codon at a time. In reality, a pre-mRNA is first chopped into smaller segments called introns and exons. For the purposes of protein translation, the introns are thrown out, and the exons are glued together sequentially to produce a final strand of mRNA. This cutting and pasting process is called splicing, and it is facilitated by a collection of RNA and proteins called a spliceosome. The fact that the spliceosome is made of RNA and proteins despite regulating the splicing of RNA to create proteins is just one manifestation of a molecular chicken-and-egg scenario that has yet to be fully resolved.
In terms of DNA, the exons deriving from a gene are collectively known as the gene's coding region.
Assignment
After identifying the exons and introns of an RNA string, we only need to delete the introns and concatenate the exons to form a new string ready for translation.
Write a function translate that takes the location of a FASTA file containing DNA strings. The first DNA string $$s$$ contained in the FASTA file is the string that needs to be transcribed and translated. The other DNA strings are substrings of $$s$$ acting as introns. The function must return the protein string resulting from transcribing and translating the exons of $$s$$.
Note: only one solution will exist for the dataset provided.
Example
In the following interactive session, we assume the FASTA file data.fna to be located in the current directory.
>>> translate('data.fna') 'MVYIADKQHVASREAYGHMFKVCA'