In genetics, a distinction is made between two types of mutations (the replacement of one nucleotide by a different nucleotide). A transition changes a purine nucleotide (two rings) into another purine (A ↔ G), or changes a pyrimidine nucleotide (one ring) into another pyrimidine (C ↔ T). All other mutations in which a purine is substituted for a pyrimidine, or vice versa, are called transversions.
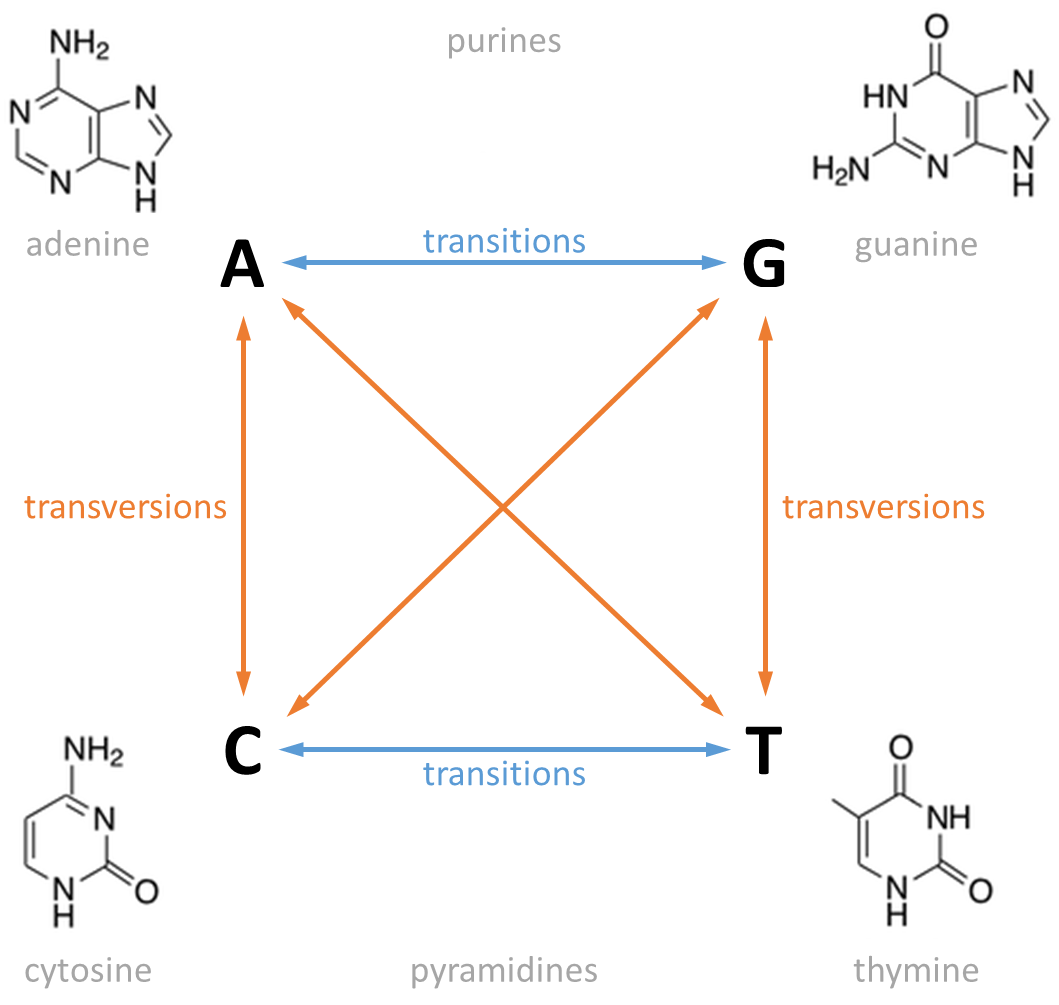
Although in theory there are only four possible transitions and eight possible transversions, in practice transitions are more likely than transversions because substituting a single ring structure for another single ring structure is more likely than substituting a double ring for a single ring. Also, transitions are less likely to result in amino acid substitutions (due to wobble base pair1) and are therefore more likely to persist as silent substitutions in populations.
Assignment
In this assignment we represent DNA sequences as strings that only contains the letters A, C, G and T. These letters represent the nucleotides that make up the DNA sequence. The nucleotides can also be represented using their lowercase variants. Given two DNA sequences $$s_1$$ and $$s_2$$ that have the same length, we define their transition/transversion ratio $$R(s_1, s_2)$$ as the ratio of the number of transitions to the number of transversions, where nucleotides at the corresponding positions between the two DNA sequences are compared with each other.
The transition/transversion ratio between homologous strands of DNA is generally about 2, but it is typically elevated in coding regions, where transversions are more likely to change the underlying amino acid and thus possibly lead to a fatal mutation in the translated protein. Point mutations that do not change this amino acid (which are thus more likely for transitions) are called silent substitutions. Your task:
-
Write a function transition that takes two nucleotides. The function must return a Boolean value that indicates whether or not replacing the first nucleotide by the second nucleotide leads to a transition.
-
Write a function transversion that takes two nucleotides. The function must return a Boolean value that indicates whether or not replacing the first nucleotide by the second nucleotide leads to a transversion.
-
Write a function ratio that takes two DNA sequences $$s_1$$ and $$s_2$$. The function may assume that both sequences have the same length (the function does not need to check this explicitly). The function must return the transition/transversion ratio $$R(s_1, s_2) \in \mathbb{R}$$ of the two given sequences as a floating point number. In case there are no transversions between the two sequences, $$R(s_1, s_2) = 0$$ by definition.
None of these function may make a distinction between uppercase and lowercase letters in the arguments passed to them.
Example
>>> transition('G', 'A')
True
>>> transition('t', 'g')
False
>>> transition('C', 'c')
False
>>> transversion('G', 'A')
False
>>> transversion('t', 'g')
True
>>> transversion('C', 'c')
False
>>> ratio('ATTAGCATTATCATC', 'AAATAGGATATATGG')
0.2222222222222222
>>> seq1 = 'GCAACGCACAACGAAAACCCTTAGGGACTGGATTATTTCGTGATCGTTGTAGTTATTGGAAGTACGGGCATCAACCCAGTT'
>>> seq2 = 'ttatctgacaaagaaagccgtcaacggctggataatttcgcgatcgtgctggttactggcggtacgagtgttcctttgggt'
>>> ratio(seq1, seq2)
1.2142857142857142