If at first you don't succeed…
We introduced the problem of finding a motif in a genetic string in "Finding a motif in DNA". More commonly, we will have a collection of motifs that we may wish to find in a larger string, for example when searching a genome for a collection of known genes.
This application sets up the algorithmic problem of pattern matching, in which we are searching a large string (called a text) for instances of a collection of smaller strings, called patterns. For the moment, we will focus on requiring that all matches should be exact.
The most obvious method for finding exact patterns in a text is to simply
apply a simple "sliding window" algorithm for each pattern. However, this
method is time-consuming if we have a large number of patterns to consider
(which will often be the case when dealing with a database of genes). It
would be better if instead of traversing the genome for every pattern, we
could somehow only traverse it once. To this end, we will need a data
structure that can efficiently organize a collection of patterns.
Assignment
Given a collection of strings, their trie (often pronounced "try" to avoid ambiguity with the general term tree) is a rooted tree formed as follows. For every unique first symbol in the strings, an edge is formed connecting the root to a new vertex. This symbol is then used to label the edge.
We may then iterate the process by moving down one level as follows. Say that an edge connecting the root to a node $$v$$ is labeled with A. Then we delete the first symbol from every string in the collection beginning with A and then treat $$v$$ as our root. We apply this process to all nodes that are adjacent to the root, and then we move down another level and continue. The figure below shows an example of a trie.
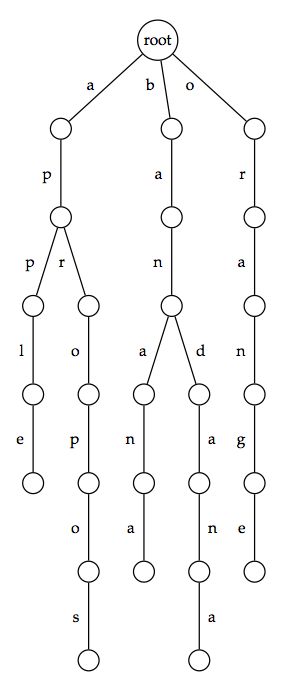
As a result of this method of construction, the symbols along the edges
of any path in the trie from the root to a leaf will spell out a unique
string from the collection, as long as no string is a prefix of another in
the collection (this would cause the first string to be encoded as a path
terminating at an internal node).
Write a function trie that takes the location of a FASTA file containing a collection of DNA strings, none of which is a prefix of another. The function must return the "adjacency list" corresponding to the trie $$T$$ for these patterns, in the following format. If $$T$$ has $$n$$ nodes, first label the root with 1 and then label the remaining nodes with the integers 2 through $$n$$ in any order you like. Each edge of $$T$$ is then encoded by a triple (a tuple containing three elements) containing the integer representing the edge's parent node, followed by the integer representing the edge's child node, and finally the symbol labeling the edge. The adjacency list return by the function is the set containing the encodings of all edges of $$T$$.
Example
In the following interactive session, we assume the FASTA file data.fna to be located in the current directory.
>>> trie('data.fna') ((1, 2, 'A'), (2, 3, 'T'), (3, 4, 'A'), (4, 5, 'G'), (5, 6, 'A'), (3, 7, 'C'), (1, 8, 'G'), (8, 9, 'A'), (9, 10, 'T'))