The genetic code
Given a nucleotide string obtained from sequencing or a database, we want to know whether this string corresponds to a coding region of the genome. If so, you need only apply the genetic code to translate your string into an amino acid chain.
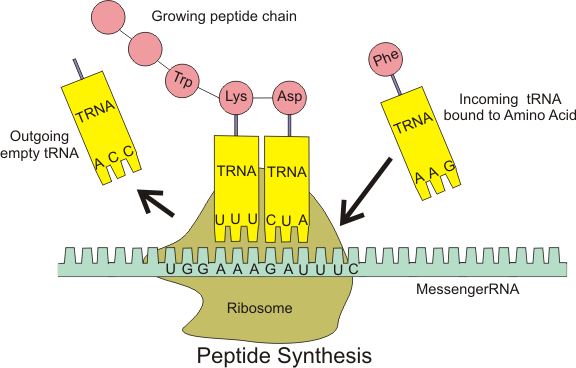
The apparent difficulty of translation is that somehow 4 RNA bases must
correspond to a protein language of 20 amino acids. In order for every
possible amino acid to be used, we must translate 3-nucleotide codons into
amino acids (see figure below). Note that there are $$4^3 = 64$$ possible
codons, so that multiple codons may encode the same amino acid. Two
special types of codons are the start codon (AUG),
which codes for the amino acid methionine and always indicates the start
of translation, and the three stop codons (UAA, UAG,
UGA), which do not code for an amino acid and terminate the
translation process.
It is important to note that some organisms and DNA-containing organelles use an alternative form of the genetic code. This phenomenon is called genetic code variation. For example, vertebrate mitochondria treat AGA and AGG as stop codons instead of having these two codons code for arginine.
Thus, it is important to check the source of your genome data prior to translation.Assignment
The 20 commonly occurring amino acids are abbreviated by using 20 letters from the English alphabet (all letters except for B, J, O, U, X, and Z). Protein strings are constructed from these 20 symbols. The RNA codon table shows the encoding from each RNA codon to the amino acid alphabet.
NCBI has a detailed list of genetic code variants (codon tables), along with indexes representing these codes (1 = standard genetic code, etc.). For now, when translating DNA and RNA strings, we will start with the first letter of the string and ignore stop codons.
Write a function geneticCodes that takes two strings: i) a DNA or RNA string $$s$$ and ii) a protein string translated from $$s$$ using of the genetic codes. The function must return a set containing the indices of all NCBI genetic code variants that translate the given DNA or RNA string into the given protein string.
Example
>>> geneticCodes('ATGGCCATGGCGCCCAGAACTGAGATCAATAGTACCCGTATTAACGGGTGA', 'MAMAPRTEINSTRING') {1, 6, 11, 12, 15, 16, 22, 23, 26} >>> from Bio.Seq import Seq >>> geneticCodes(Seq('GACTTGTAGTAAATCTATGGTCCCATCACATATGCGGAGAAC'), 'DLQIYGPITYAEN') {15}
Programming shortcut
BioPython possesses a translate() method for converting RNA strings to protein strings:
translate(sequence, table='Standard', stop_symbol='*', to_stop=False)
The translate() method has the following parameters:
- sequence: the DNA or RNA string to translate
- table: the codon table to use; this can be either a name (string) or NCBI identifier (integer); defaults to the "Standard" table, which has index 1
- stop_symbol: a single symbol used to mark any terminators, which defaults to the asterisk (*)
- to_stop: a Boolean value; if True, translation is terminated at the first stop codon appearing in the frame; defaults to False
Here are some examples of translate() in action:
>>> from Bio.Seq import translate >>> coding_dna = 'GTGGCCATTGTAATGGGCCGCTGAAAGGGTGCCCGATAG' >>> translate(coding_dna) 'VAIVMGR*KGAR*' >>> translate(coding_dna, stop_symbol='@') 'VAIVMGR@KGAR@' >>> translate(coding_dna, to_stop=True) 'VAIVMGR' >>> translate(coding_dna, table=2) 'VAIVMGRWKGAR*' >>> translate(coding_dna, table=2, to_stop=True) 'VAIVMGRWKGAR'