The workhorse of peptide sequencing is the mass spectrometer, an expensive molecular scale that shatters molecules into pieces and then weighs the resulting fragments. The mass spectrometer measures the mass of a molecule in daltons (Da), where 1 Da is approximately equal to the mass of a single nuclear particle (i.e., a proton or neutron).
We will approximate the mass of a molecule by simply adding the number of protons and neutrons found in the molecule's constituent atoms, which yields the molecule's integer mass. For example, the amino acid Gly — which has chemical formula C2H3ON — has an integer mass of 57, since \[2 \times 12 + 3 \times 1 + 1 \times 16 + 1 \times 14 = 57\] Yet 1 Da is not exactly equal to the mass of a proton/neutron, and we may need to account for different naturally occurring isotopes of each atom when weighing a molecule. As a result, amino acids typically have non-integer masses (e.g., Gly has total mass equal to approximately 57.02 Da). For simplicity, however, we will work with the following integer mass table.
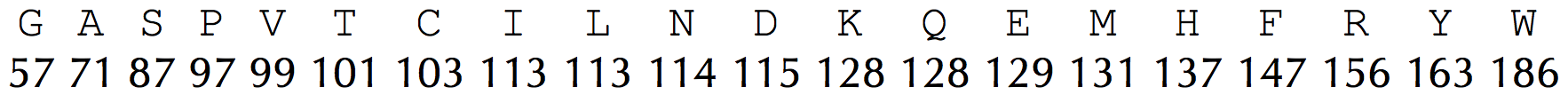
The theoretical spectrum of a cyclic peptide $$p$$ — denoted Cyclospectrum($$p$$) — is the collection of all of the masses of its subpeptides, in addition to the mass 0 and the mass of the entire peptide. We will assume that the theoretical spectrum can contain duplicate elements, as is the case for NQEL (shown below), where NQ and EL have the same mass.

Assignment
Write a function cyclic_spectrum that takes a peptide $$p$$. The function must return the theoretical spectrum of the cyclic peptide $$p$$ (i.e. Cyclospectrum($$p$$)).
Example
>>> cyclic_spectrum('LEQN') (0, 113, 114, 128, 129, 227, 242, 242, 257, 355, 356, 370, 371, 484)