Transcription may begin anywhere
In "Transcribing DNA into RNA", we discussed the transcription of DNA into RNA, and in "Translating RNA into protein", we examined the translation of RNA into a chain of amino acids for the construction of proteins. We can view these two processes as a single step in which we directly translate a DNA string into a protein string, thus calling for a DNA codon table.
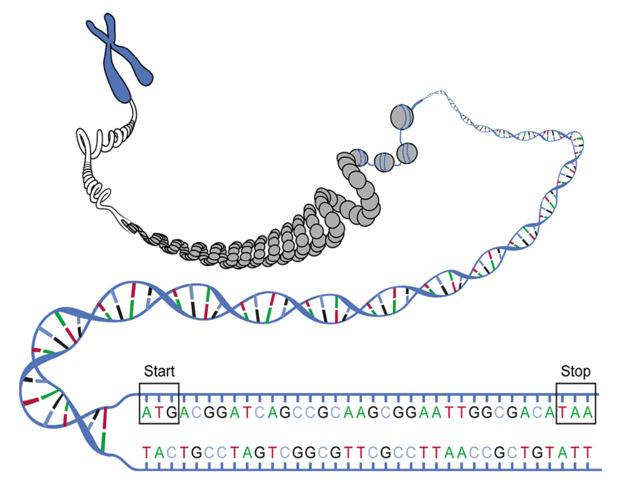
However, three immediate wrinkles of complexity arise when we try to pass directly from DNA to proteins. First, not all DNA will be transcribed into RNA: so-called junk DNA appears to have no practical purpose for cellular function. Second, we can begin translation at any position along a strand of RNA, meaning that any substring of a DNA string can serve as a template for translation, as long as it begins with a start codon, ends with a stop codon, and has no other stop codons in the middle (see figure above). As a result, the same RNA string can actually be translated in three different ways, depending on how we group triplets of symbols into codons. For example, …AUGCUGAC… can be translated as …AUGCUG…, …UGCUGA…, and …GCUGAC…, which will typically produce wildly different protein strings.
Assignment
Either strand of a DNA double helix can serve as the coding strand for RNA transcription. Hence, a given DNA string implies six total reading frames, or ways in which the same region of DNA can be translated into amino acids: three reading frames result from reading the string itself, whereas three more result from reading its reverse complement.
An open reading frame (ORF) is one which starts from the start codon and ends by a stop codon, without any other stop codons in between. Thus, a candidate protein string is derived by translating an open reading frame into amino acids until a stop codon is reached.
Write a function candidateTranslations that takes a DNA string $$s$$. The function must return a set containing every distinct candidate protein string that can be translated from ORFs of $$s$$.
Example
>>> ORFs('AGCCATGTAGCTAACTCAGGTTACATGGGGATGACCCCGCGACTTGGATTAGAGTCTCTTTTGGAATAAGCCTGAATGATCCGAGTAGCATCTCAG') {'M', 'MTPRLGLESLLE', 'MGMTPRLGLESLLE', 'MLLGSFRLIPKETLIQVAGSSPCNLS'} >>> from Bio import SeqIO >>> translate(*SeqIO.parse('data.fna', 'fasta')) {'MACTGGREYYLFTRCPPRARISDALSTATHPSVASRWPRF', 'MGLSVRIDPLRNQTAGSGCQPITSDNRLIRMLGQFRIVV', 'MTKMCNKEFPQYMSPNC', 'MGDPVTRLTTS', 'MHRWV', 'MSSPGPDF', 'MKRLKDACI', 'MCNKEFPQYMSPNC', 'MFSRIFCAWPGADYTLKVAYR', 'MVRSAMWR', 'MRN', 'MPWVTLSPA', 'MRIRAAKLCNPSDWEKPSSRLAAM', 'MATPDFVRLEDLLNCCNSL', 'MWR', 'MQL', 'MRDNRVSLLLLPLLTDGFTIRGDARSVPHSSLAKLLCGTDRAFVSACYH', 'M', 'MSPNC', 'MHDENV', 'MLGQFRIVV', 'MGLYWYARQQGITAIITPTY', 'MYRRKGILPVYQMSSPGPDF'}