We define the convolution of a cyclic spectrum by taking all positive differences of masses in the spectrum. The figure below shows the convolution of the theoretical spectrum of “NQEL”.
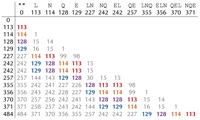
Some of the values appear more frequently than others. For example, 113 (the mass of “L”) appears eight times in the convolution of the theoretical spectrum of “NQEL”; we say that 113 has multiplicity 8. Six of the eight occurrences of 113 correspond to subpeptide pairs differing in an “L”: “L” and “”; “LN” and “N”; “EL” and “E”; “LNQ” and “NQ”; “QEL” and “QE”; “NQEL” and “NQE”.
The algorithm ConvolutionCyclopeptideSequencing first computes the convolution a given experimental spectrum. It then selects the M most frequent elements (with ties) between 57 and 200 in the convolution to form an extended alphabet of amino acid masses. Finally it runs the algorithm LeaderboardCyclopeptideSequencing, where amino acids are restricted to this alphabet.
Assignment
Write a Python function leaderpeptide that implements the above algorithm ConvolutionCyclopeptideSequencing. The functions takes as parameters an integer M, an integer N, and a collection of (possibly repeated) integers Spectrum. It returns a cyclic peptide with amino acids taken only from the top M elements (and ties) of the convolution of the given spectrum that fall between 57 and 200, and where the size of the the leaderboard is restricted to the top N (and ties).
Example
>>> leaderpeptide(20, 60, [57, 57, 71, 99, 129, 137, 170, 186, 194, 208, 228, 265, 285, 299, 307, 323, 356, 364, 394, 422, 493])
'99-71-137-57-72-57'