Much like replication, the chemical machinery underlying transcription and translation is fascinating, but from a computational perspective, both processes are straightforward. Transcription simply transforms a DNA string into an RNA string by replacing all occurrences of T with U. The resulting strand of RNA is translated into an amino acid sequence via the genetic code. This process converts each 3-mer of RNA — called a codon — into one of 20 amino acids.
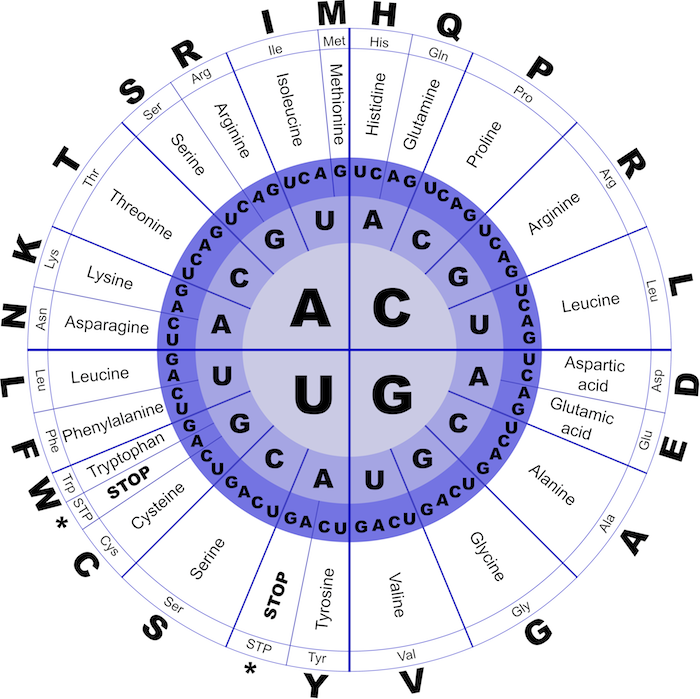
As illustrated in the figure above, each of the 64 RNA codons encodes its own amino acid (some codons encode the same amino acid), with the exception of three stop codons that do not translate into amino acids and serve to halt translation. For example, the DNA string TATACGAAA transcribes into the RNA string UAUACGAAA, which in turn translates into the amino acid string Tyr-Thr-Lys.
The following problem asks you to find the translation of an RNA string into an amino acid string.
Assignment
Write a function translate that takes the locations of two FASTA files. The first FASTA file contains an RNA string $$s$$ corresponding to a strand of mRNA. The function must write the translation of $$s$$ into an amino acid string to the FASTA file whose location is passed as the second argument. Stop codons must be represented by an asterisk (*).Example
In the following interactive session, we assume the FASTA files data01.fna, data02.fna, data01.faa and data02.faa to be located in the current directory.
>>> translate('data01.fna', 'data01.faa') >>> print(open('data01.faa').read().rstrip()) >seq01 MAMAPRTEINSTRING* >>> translate('data02.fna', 'data02.faa') >>> print(open('data02.faa').read().rstrip()) >seq01 MYHDISRSIMEANNTMKSEHGPYIWLGLWGWFFFNNSPTMIFELNWHIQFLW*