The genetic code describes how the information in the genetic material (DNA1 or RNA2) of life's cells is read to form a protein3. The genetic code was unraveled in the 1960s, a few years after the discovery of the DNA helix. It explains how the four-letter code of DNA (represented by the letters A, C, G and T) can be translated4 into the 20 different amino acids5 (represented by 20 different letters), the building blocks of proteins. With few exceptions, the genetic code is universally valid in all forms of life.
Fragments of DNA encoding proteins (genes6) are transferred in living cells to a single-stranded RNA molecule. This molecule — the messenger RNA7 — is essentially a copy of DNA and carries instructions for making a protein. In the ribosome8, amino acids are linked together in an order determined by the nucleotide sequence in the mRNA. In the process, the ribosome reads three nucleotides from the mRNA each time, each corresponding to a specific amino acid. This process is called translation9.
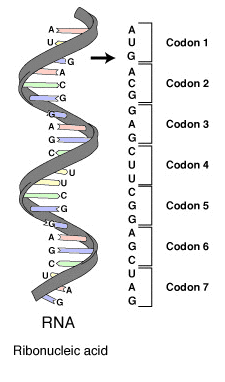
The three consecutive nucleotides are called a codon10. Codons specify which amino acids are added to the protein chain, and also determine the start and stop signals of the translation process. Usually, the term codon refers to the nucleotides found in messenger RNA: adenine (A), cytosine (C), guanine (G) and uracil (U). For example, the codon ACU corresponds to the amino acid threonine11, and CGG to arginine12. The genetic code can be summarised in a simple codon table of 64 codes.
It has long been assumed that the genetic code is universal — a given codon would code for the same amino acid in every organism. This assumption is largely true: the genetic code applies in all three domains19 of life. This fact provides an important argument for the common descent20 of all life forms. However, a few exceptions — variations — to the genetic code have been discovered. For example, the unicellular fungus Candida albicans21 translates the codon CUG to serine22, while almost all other organisms translate this codon to leucine23. In some ciliates24 (single-celled eukaryotes), the three conventional stop codons work differently — namely, these simply code for amino acids, and the end of the translation is signalled by the 3'-end of the mRNA.
In mitochondria25 of diverse species, genetic codes also vary widely. In mammalian mitochondria, for example, the codon AUA is translated to methionine26, instead of AUG. Mitochondria have their own genetic material and encode their own translational machinery. Such variations show that the genetic code is not unshakably fixed, but can undergo evolutionary changes.
Assignment
The DNA alphabet consists of four different nucleotides represented by the letters A, C, G and T. A DNA sequence (str) consists of a sequence of letters from the DNA alphabet. The RNA alphabet consists of four different nucleotides represented by the letters A, C, G and U. An RNA sequence (str) consists of a sequence of letters from the RNA alphabet. When transcribing DNA to RNA, thymine (T) is converted to uracil (U), making the letter T in the DNA alphabet a synonym for the letter U in the RNA alphabet. A codon (str) is a sequence (DNA or RNA) of length 3.
The protein alphabet consists of 20 different amino acids (represented by 20 different letters) and a stop codon (represented by an asterisk: *). A protein sequence (str) consists of a sequence of characters from the protein alphabet.
A DNA or RNA sequence is translated into a protein sequence by converting each codon of three consecutive letters into the corresponding amino acid or stop codon. If the length of the DNA or RNA sequence is not a multiple of three, the last letter or the last two letters of the sequence are ignored in the translation. The genetic code used for that translation is recorded in a translation table: a text file consisting of 64 lines. Each line contains a unique codon and the corresponding amino acid, separated by a space. For the codons, the translation table uses either the DNA alphabet (with T for thymine) or the RNA alphabet (with U for uracil).
Define a class GeneticCode that allows representing genetic codes. When creating a genetic code (GeneticCode), the location (str) of the translation table of the genetic code must be passed. On a genetic code $$\mathcal{C}$$ (GeneticCode), you must be able to call at least the following methods:
-
A method amino_acid that takes a codon $$c$$ (str; DNA or RNA). If $$c$$ does not represent a valid codon, an AssertionError must be raised with the message invalid codon. Otherwise, the amino acid (str; uppercase) corresponding to codon $$c$$ according to genetic code $$\mathcal{C}$$ must be returned. When translating codon $$c$$, no distinction should be made between uppercase and lowercase letters, nor between the letters U (uracil) and T (thymine). As a result, both DNA and RNA codons can be passed to the method.
-
A method protein that takes a sequence $$s$$ (str; DNA or RNA). This sequence may contain either uppercase or lowercase letters. If $$s$$ does not represent a valid DNA or RNA sequence, an AssertionError must be raised with the message invalid sequence. Otherwise, the protein sequence (str; in uppercase) obtained by translating sequence $$s$$ according to genetic code $$\mathcal{C}$$ must be returned.
Example
In the following interactive session, we assume the current directory contains the text file standard_code.txt27. This file contains the translation table of the standard genetic code28, where the specification of codons uses the DNA alphabet.
>>> code = GeneticCode('standard_code.txt29')
>>> code.amino_acid('AGT')
'S'
>>> code.amino_acid('cga')
'R'
>>> code.amino_acid('UCU')
'S'
>>> code.amino_acid('ABC')
Traceback (most recent call last):
AssertionError: invalid codon
>>> code.amino_acid('aagc')
Traceback (most recent call last):
AssertionError: invalid codon
>>> code.protein('ATGCTGATGATGGGCTATTATCGAT')
'MLMMGYYR'
>>> code.protein('uauccuaguguc')
'YPSV'
>>> code.protein('AAGTCGTAGCTACGXXXXGAGAAGGAT')
Traceback (most recent call last):
AssertionError: invalid sequence